Metagenomic source tracking after microbiota transplant therapy
- PMID: 40229213
- PMCID: PMC12005403
- DOI: 10.1080/19490976.2025.2487840
Metagenomic source tracking after microbiota transplant therapy
Abstract
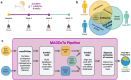
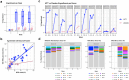
Reliable engraftment assessment of donor microbial communities and individual strains is an essential component of characterizing the pharmacokinetics of microbiota transplant therapies (MTTs). Recent methods for measuring donor engraftment use whole-genome sequencing and reference databases or metagenome-assembled genomes (MAGs) to track individual bacterial strains but lack the ability to disambiguate DNA that matches both donor and patient microbiota. Here, we describe a new, cost-efficient analytic pipeline, MAGEnTa, which compares post-MTT samples to a database comprised MAGs derived directly from donor and pre-treatment metagenomic data, without relying on an external database. The pipeline uses Bayesian statistics to determine the likely sources of ambiguous reads that align with both the donor and pre-treatment samples. MAGEnTa recovers engrafted strains with minimal type II error in a simulated dataset and is robust to shallow sequencing depths in a downsampled dataset. Applying MAGEnTa to a dataset from a recent MTT clinical trial for ulcerative colitis, we found the results to be consistent with 16S rRNA gene SourceTracker analysis but with added MAG-level specificity. MAGEnTa is a powerful tool to study community and strain engraftment dynamics in the development of MTT-based treatments that can be integrated into frameworks for functional and taxonomic analysis.
Keywords: Metagenomics; fecal microbiota transplant; metagenome-assembled genome; microbiota engraftment; microbiota transplant therapy; shotgun sequencing; ulcerative colitis; whole-genome sequencing.
Conflict of interest statement
AK has patents pertaining to the purification and lyopreservation of fecal microbiota for microbiota transplant therapy. Byron Vaughn: Other research support not for this project from: Takeda, KateFarms, Diasorin, Nestle, Roche. Consulting for HealthDelegates. The other authors do not have any disclosures.
Figures

References
-
- Khanna S, Assi M, Lee C, Yoho D, Louie T, Knapple W, Aguilar H, Garcia-Diaz J, Wang GP, Berry SM, et al. Efficacy and safety of RBX2660 in PUNCH CD3, a phase III, randomized, double-blind, placebo-controlled trial with a bayesian primary analysis for the prevention of recurrent clostridioides difficile infection. Drugs. 2022;82(15):1527–13. doi: 10.1007/s40265-022-01797-x. - DOI - PMC - PubMed
-
- Podlesny D, Durdevic M, Paramsothy S, Kaakoush NO, Högenauer C, Gorkiewicz G, Walter J, Fricke WF.. Identification of clinical and ecological determinants of strain engraftment after fecal microbiota transplantation using metagenomics. Cell Rep Med. 2022;3(8):100711. doi: 10.1016/j.xcrm.2022.100711. - DOI - PMC - PubMed
-
- Kang DW, Adams JB, Gregory AC, Borody T, Chittick L, Fasano A, Khoruts A, Geis E, Maldonado J, McDonough-Means S, et al. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome. 2017;5(1):10. doi: 10.1186/s40168-016-0225-7. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
