A genetically informed brain atlas for enhancing brain imaging genomics
- PMID: 40229250
- PMCID: PMC11997130
- DOI: 10.1038/s41467-025-57636-6
A genetically informed brain atlas for enhancing brain imaging genomics
Abstract
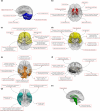
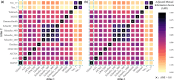
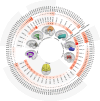
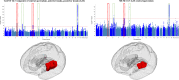
Brain imaging genomics has manifested considerable potential in illuminating the genetic determinants of human brain structure and function. This has propelled us to develop the GIANT (Genetically Informed brAiN aTlas) that accounts for genetic and neuroanatomical variations simultaneously. Integrating voxel-wise heritability and spatial proximity, GIANT clusters brain voxels into genetically informed regions, while retaining fundamental anatomical knowledge. Compared to conventional (non-genetics) brain atlases, GIANT exhibits smaller intra-region variations and larger inter-region variations in terms of voxel-wise heritability. As a result, GIANT yields increased regional SNP heritability, enhanced polygenicity, and its polygenic risk score explains more brain volumetric variation than traditional neuroanatomical brain atlases. We provide extensive validation to GIANT and demonstrate its neuroanatomical validity, confirming its generalizability across populations with diverse genetic ancestries and various brain conditions. Furthermore, we present a comprehensive genetic architecture of the GIANT regions, covering their functional annotation at the molecular levels, their associations with other complex traits/diseases, and the genetic and phenotypic correlations among GIANT-defined imaging endophenotypes. In summary, GIANT constitutes a brain atlas that captures the complexity of genetic and neuroanatomical heterogeneity, thereby enhancing the discovery power and applicability of imaging genomics investigations in biomedical science.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

