Specific labeling of outer membrane vesicles with antibiotic-conjugated probe reveals early bacterial infections in blood
- PMID: 40229269
- PMCID: PMC11997070
- DOI: 10.1038/s41467-025-58676-8
Specific labeling of outer membrane vesicles with antibiotic-conjugated probe reveals early bacterial infections in blood
Abstract
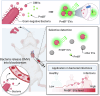
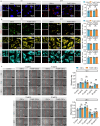
Bacterial outer membrane vesicles (OMVs) are nano-sized structures derived from the outer membrane of Gram-negative bacteria, which have emerged as key players in host-pathogen interactions, yet their potential as biomarkers remains largely unexplored due to the difficulty of identification in complex biological samples. Here we show an approach for detecting and quantifying bacterial OMVs in blood using a Polymyxin B-fluorescein probe (PmBF), which targets bacterial lipopolysaccharides (LPS). The probe selectively labels OMVs, enabling their differentiation from host extracellular vesicles and quantitative analysis using nano-flow cytometry. In male mouse models of pneumonia, we observe elevated serum PmBF+ EVs as early as 6 h post-infection, preceding positive blood cultures. In clinical samples, PmBF+ EVs show superior performance for diagnosing bacterial infections and differentiate them from virus or mycoplasma infections. Our findings highlight circulating PmBF+ EVs as promising biomarkers of bacterial infections.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Benzimidazole-Derived B2 as a Fluorescent Probe for Bacterial Outer Membrane Vesicle (OMV) Labeling: Integrating DFT, Molecular Dynamics, Flow Cytometry, and Confocal Microscopy.Int J Mol Sci. 2025 May 14;26(10):4682. doi: 10.3390/ijms26104682. Int J Mol Sci. 2025. PMID: 40429826 Free PMC article.
-
Lipid A in outer membrane vesicles shields bacteria from polymyxins.J Extracell Vesicles. 2024 May;13(5):e12447. doi: 10.1002/jev2.12447. J Extracell Vesicles. 2024. PMID: 38766978 Free PMC article.
-
Extracellular RNAs in periodontopathogenic outer membrane vesicles promote TNF-α production in human macrophages and cross the blood-brain barrier in mice.FASEB J. 2019 Dec;33(12):13412-13422. doi: 10.1096/fj.201901575R. Epub 2019 Sep 21. FASEB J. 2019. PMID: 31545910 Free PMC article.
-
Bacterial Outer Membrane Vesicles as Antibiotic Delivery Vehicles.Front Immunol. 2021 Sep 20;12:733064. doi: 10.3389/fimmu.2021.733064. eCollection 2021. Front Immunol. 2021. PMID: 34616401 Free PMC article. Review.
-
Bacterial Membrane Vesicles in Pneumonia: From Mediators of Virulence to Innovative Vaccine Candidates.Int J Mol Sci. 2021 Apr 8;22(8):3858. doi: 10.3390/ijms22083858. Int J Mol Sci. 2021. PMID: 33917862 Free PMC article. Review.
Cited by
-
Adaptations of Bacterial Extracellular Vesicles in Response to Antibiotic Pressure.Int J Mol Sci. 2025 May 23;26(11):5025. doi: 10.3390/ijms26115025. Int J Mol Sci. 2025. PMID: 40507835 Free PMC article. Review.
References
-
- World Health Organization, WHO bacterial priority pathogens list, 2024: Bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance (https://www.who.int/publications/i/item/9789240093461).
MeSH terms
Substances
Grants and funding
- 82302593/National Natural Science Foundation of China (National Science Foundation of China)
- 82272438/National Natural Science Foundation of China (National Science Foundation of China)
- 2023A1515012512/Natural Science Foundation of Guangdong Province (Guangdong Natural Science Foundation)
- 2023B1515020058/Natural Science Foundation of Guangdong Province (Guangdong Natural Science Foundation)
- 202301-202/Guangzhou Medical University (Guangzhou Medical College)
LinkOut - more resources
Full Text Sources
Medical

