PIN1-SUMO2/3 motif suppresses excessive RNF168 chromatin accumulation and ubiquitin signaling to promote IR resistance
- PMID: 40229270
- PMCID: PMC11997057
- DOI: 10.1038/s41467-025-56974-9
PIN1-SUMO2/3 motif suppresses excessive RNF168 chromatin accumulation and ubiquitin signaling to promote IR resistance
Abstract
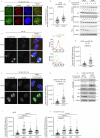
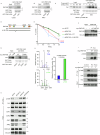
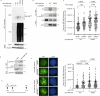
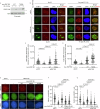
RNF168 is an E3 ubiquitin ligase critical to the mammalian DNA double-strand break repair response. The protein is recruited to and amplifies ubiquitin signals at damaged chromatin and, if not properly regulated, can drive an uncontrolled ubiquitin cascade potentially harmful to repair outcomes. Several indirect mechanisms restrict RNF168 positive feedback, and a longstanding question has been whether these alone suppress excessive RNF168 signaling or whether mechanisms to remove RNF168 from damaged chromatin exist. Here, we reveal a cascade of post-translational modifications which act at three adjacent amino acids, threonine-208, proline-209 and lysine-210, to process RNF168 actively. Phosphorylation at threonine-208 by CDK1/2 induces interaction with the peptidyl-prolyl isomerase PIN1. PIN1 promotes RNF168 SUMOylation at lysine-210, resulting in p97/VCP mediated removal. These actions promote RNF168 clearance and limit RNF168 chromatin build-up. Thus, single amino acid substitutions of the regulatory motif (SUMO-PIN1-assisted Chromatin Regulator, SPaCR) that restrict PIN1 interaction or SUMOylation are sufficient to drive supraphysiological accumulation of RNF168, increased ubiquitin signaling, excessive 53BP1 recruitment and radiosensitivity. Our findings define a mechanism of direct RNF168 regulation that is part of the normal damage response, promoting RNF168 dissociation from chromatin and limiting deleterious ubiquitin signaling.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Doil, C. et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell136, 435–446 (2009). - PubMed
-
- Mailand, N. et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell131, 887–900 (2007). - PubMed
-
- Stewart, G. S. et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell136, 420–434 (2009). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

