Programmable protein stabilization with language model-derived peptide guides
- PMID: 40229275
- PMCID: PMC11997201
- DOI: 10.1038/s41467-025-58872-6
Programmable protein stabilization with language model-derived peptide guides
Abstract
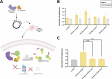
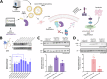
Dysregulated protein degradation via the ubiquitin-proteasomal pathway can induce numerous disease phenotypes, including cancer, neurodegeneration, and diabetes. While small molecule-based targeted protein degradation (TPD) and targeted protein stabilization (TPS) platforms can address this dysregulation, they rely on structured and stable binding pockets, which do not exist to classically "undruggable" targets. Here, we expand the TPS target space by engineering "deubiquibodies" (duAbs) via fusion of computationally-designed peptide binders to the catalytic domain of the potent OTUB1 deubiquitinase. In human cells, duAbs effectively stabilize exogenous and endogenous proteins in a DUB-dependent manner. Using protein language models to generate target-binding peptides, we engineer duAbs to conformationally diverse target proteins, including key tumor suppressor proteins p53 and WEE1, and heavily-disordered fusion oncoproteins, such as PAX3::FOXO1. We further encapsulate p53-targeting duAbs as mRNA in lipid nanoparticles and demonstrate effective intracellular delivery, p53 stabilization, and apoptosis activation, motivating further in vivo translation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: P.C., L.H., and M.P.D. are listed as inventors on US Patent Application 63/541,921: “Peptide-Guided Protein Stabilizers and Uses Thereof”. P.C. and M.P.D. are co-founders of UbiquiTx, Inc., which commercializes genetically encoded proteome editing technologies, and are co-inventors of duAb patents. P.C.’s interests are reviewed and managed by Duke University in accordance with their conflict-of-interest policies. M.P.D.’s interests are reviewed and managed by Cornell University in accordance with their conflict-of-interest policies.
Figures


Update of
-
Programmable Protein Stabilization with Language Model-Derived Peptide Guides.Res Sq [Preprint]. 2024 Jul 26:rs.3.rs-4670386. doi: 10.21203/rs.3.rs-4670386/v1. Res Sq. 2024. Update in: Nat Commun. 2025 Apr 15;16(1):3555. doi: 10.1038/s41467-025-58872-6. PMID: 39108486 Free PMC article. Updated. Preprint.
Similar articles
-
Programmable Protein Stabilization with Language Model-Derived Peptide Guides.Res Sq [Preprint]. 2024 Jul 26:rs.3.rs-4670386. doi: 10.21203/rs.3.rs-4670386/v1. Res Sq. 2024. Update in: Nat Commun. 2025 Apr 15;16(1):3555. doi: 10.1038/s41467-025-58872-6. PMID: 39108486 Free PMC article. Updated. Preprint.
-
YTHDF2 protein stabilization by the deubiquitinase OTUB1 promotes prostate cancer cell proliferation via PRSS8 mRNA degradation.J Biol Chem. 2024 Apr;300(4):107152. doi: 10.1016/j.jbc.2024.107152. Epub 2024 Mar 9. J Biol Chem. 2024. PMID: 38462165 Free PMC article.
-
FOXO1 promotes cancer cell growth through MDM2-mediated p53 degradation.J Biol Chem. 2024 Apr;300(4):107209. doi: 10.1016/j.jbc.2024.107209. Epub 2024 Mar 21. J Biol Chem. 2024. PMID: 38519029 Free PMC article.
-
p53 stability is regulated by diverse deubiquitinating enzymes.Biochim Biophys Acta Rev Cancer. 2017 Dec;1868(2):404-411. doi: 10.1016/j.bbcan.2017.08.001. Epub 2017 Aug 8. Biochim Biophys Acta Rev Cancer. 2017. PMID: 28801249 Review.
-
Otubain 1: a non-canonical deubiquitinase with an emerging role in cancer.Endocr Relat Cancer. 2019 Jan 1;26(1):R1-R14. doi: 10.1530/ERC-18-0264. Endocr Relat Cancer. 2019. PMID: 30400005 Free PMC article. Review.
Cited by
-
Target sequence-conditioned design of peptide binders using masked language modeling.Nat Biotechnol. 2025 Aug 13. doi: 10.1038/s41587-025-02761-2. Online ahead of print. Nat Biotechnol. 2025. PMID: 40804173
-
PTM-Mamba: a PTM-aware protein language model with bidirectional gated Mamba blocks.Nat Methods. 2025 May;22(5):945-949. doi: 10.1038/s41592-025-02656-9. Epub 2025 Apr 10. Nat Methods. 2025. PMID: 40211004 Free PMC article.
References
-
- Sabapathy, K. & Lane, D. P. Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat. Rev. Clin. Oncol.15, 13–30 (2018). - PubMed
-
- Ward, C. L., Omura, S. & Kopito, R. R. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell83, 121–127 (1995). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

