H3.1K27M-induced misregulation of the TONSOKU-H3.1 pathway causes genomic instability
- PMID: 40229276
- PMCID: PMC11997104
- DOI: 10.1038/s41467-025-58892-2
H3.1K27M-induced misregulation of the TONSOKU-H3.1 pathway causes genomic instability
Abstract
The oncomutation lysine 27-to-methionine in histone H3 (H3K27M) is frequently identified in tumors of patients with diffuse midline glioma-H3K27 altered (DMG-H3K27a). H3K27M inhibits the deposition of the histone mark H3K27me3, which affects the maintenance of transcriptional programs and cell identity. Cells expressing H3K27M are also characterized by defects in genome integrity, but the mechanisms linking expression of the oncohistone to DNA damage remain mostly unknown. In this study, we demonstrate that expression of H3.1K27M in the model plant Arabidopsis thaliana interferes with post-replicative chromatin maturation mediated by the H3.1K27 methyltransferases ATXR5 and ATXR6. As a result, H3.1 variants on nascent chromatin remain unmethylated at K27 (H3.1K27me0), leading to ectopic activity of TONSOKU (TSK/TONSL), which induces DNA damage and genomic alterations. Elimination of TSK activity suppresses the genome stability defects associated with H3.1K27M expression, while inactivation of specific DNA repair pathways prevents survival of H3.1K27M-expressing plants. Overall, our results suggest that H3.1K27M disrupts the chromatin-based mechanisms regulating TSK activity, which causes genomic instability and may contribute to the etiology of DMG-H3K27a.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

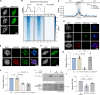

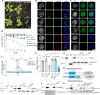

Update of
-
H3.1K27M-induced misregulation of the TSK/TONSL-H3.1 pathway causes genomic instability.bioRxiv [Preprint]. 2024 Dec 11:2024.12.09.627617. doi: 10.1101/2024.12.09.627617. bioRxiv. 2024. Update in: Nat Commun. 2025 Apr 14;16(1):3547. doi: 10.1038/s41467-025-58892-2. PMID: 39713323 Free PMC article. Updated. Preprint.
References
-
- Louis, D. N. et al. cIMPACT-NOW update 2: diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathol.135, 639–642 (2018). - PubMed
-
- Schwartzentruber, J. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature482, 226–231 (2012). - PubMed
-
- Bender, S. et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell24, 660–672 (2013). - PubMed
MeSH terms
Substances
Grants and funding
- R01 AG078926/AG/NIA NIH HHS/United States
- 104175/Z/14/Z/Wellcome Trust (Wellcome)
- P01 CA196539/CA/NCI NIH HHS/United States
- R01 AI165840/AI/NIAID NIH HHS/United States
- R35 GM128661/GM/NIGMS NIH HHS/United States
- CA196539/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- P30 CA016359/CA/NCI NIH HHS/United States
- 1S10OD030363-01A1/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- P30CA016359/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R35GM128661/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01AG078926/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- WT_/Wellcome Trust/United Kingdom
- S10 OD030363/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

