Targeting site-specific N-glycosylated B7H3 induces potent antitumor immunity
- PMID: 40229277
- PMCID: PMC11997214
- DOI: 10.1038/s41467-025-58740-3
Targeting site-specific N-glycosylated B7H3 induces potent antitumor immunity
Abstract
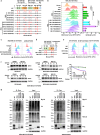
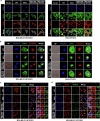
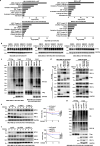
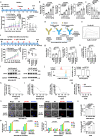
B7H3, an immune checkpoint molecule, is a highly N-glycosylated membrane protein. However, the key glycosylated asparagine residues that mediate the function of the B7H3 protein are still unclear. Here we identify that N-glycans attached to asparagine residues N91/309 and N104/322 are required for proper B7H3 localization on the cell surface membrane. We demonstrate that mutations in these two pairs of N-glycosylation sites induce ER accumulation of B7H3 by blocking its ER-to-Golgi translocation and subsequently promote its degradation via the endoplasmic reticulum-associated protein degradation pathway. Additional evidence suggests that N-glycosylation at N91/309 and N104/322 of B7H3 is essential for its inhibition of T-cell proliferation and activation. More importantly, a monoclonal antibody, Ab-82, preferentially targeting B7H3 glycosylated at N91/309 and N104/322 is developed, which exhibits the ability to elicit cytotoxic T lymphocyte-mediated antitumor immunity via B7H3 internalization. Together, these findings offer a rationale for targeting glycosylated B7H3 as a potential strategy for immunotherapy.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures



References
-
- Pinho, S. S. & Reis, C. A. Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer15, 540–555 (2015). - PubMed
-
- de Caestecker, C. & Macara, I. G. A size filter at the Golgi regulates apical membrane protein sorting. Nat. cell Biol.26, 1678–1690 (2024). - PubMed
-
- Fang, J. et al. Glucose-mediated N-glycosylation of RPTPalpha affects its subcellular localization and Src activation. Oncogene42, 1058–1071 (2023). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

