Group A Streptococcal asparagine metabolism regulates bacterial virulence
- PMID: 40229432
- PMCID: PMC12117059
- DOI: 10.1038/s44319-025-00447-z
Group A Streptococcal asparagine metabolism regulates bacterial virulence
Abstract
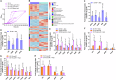
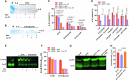
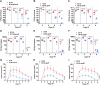
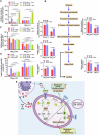
Group A Streptococcus (GAS) causes various human diseases linked to virulome expression predominantly regulated by the two-component system (TCS), CovR/S. Here, we demonstrate that asparagine (Asn) presence in a minimal chemically defined medium increases virulence gene expression in a CovR-dependent fashion. It also decreases the transcription of asparagine synthetase (AsnA), the ABC transporter responsible for Asn uptake (GlnPQ), and that of the hemolysin toxins responsible for scavenging Asn from the host. Metabolomics data show that Asn availability increases intracellular ADP/ATP ratio, which enhances phosphatase activity in structurally related CovS sensors and is probably responsible for the Asn-mediated decrease in CovR phosphorylation. Mutants deficient in AsnA, GlnPQ, asparaginase, (AsnB) activities are attenuated in a mouse model of human GAS invasive soft tissue infection. The similarity between the mechanisms of Asn-mediated regulation of GAS virulence and tumor growth suggests that, as in cancer, components maintaining Asn homeostasis could be targeted for anti-GAS treatments.
Keywords: Asparagine; Group A Streptococcus; Metabolism; Regulation Mechanism; Virulence.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures


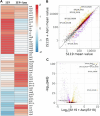
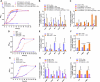




Similar articles
-
RocA Regulates Phosphatase Activity of Virulence Sensor CovS of Group A Streptococcus in Growth Phase- and pH-Dependent Manners.mSphere. 2020 May 20;5(3):e00361-20. doi: 10.1128/mSphere.00361-20. mSphere. 2020. PMID: 32434842 Free PMC article.
-
An extracellular bacterial pathogen modulates host metabolism to regulate its own sensing and proliferation.Cell. 2014 Jan 16;156(1-2):97-108. doi: 10.1016/j.cell.2013.12.007. Cell. 2014. PMID: 24439371 Free PMC article.
-
Complement-mediated opsonization of invasive group A Streptococcus pyogenes strain AP53 is regulated by the bacterial two-component cluster of virulence responder/sensor (CovRS) system.J Biol Chem. 2013 Sep 20;288(38):27494-27504. doi: 10.1074/jbc.M113.494864. Epub 2013 Aug 8. J Biol Chem. 2013. PMID: 23928307 Free PMC article. Clinical Trial.
-
Induction of endoplasmic reticulum stress and unfolded protein response constitutes a pathogenic strategy of group A streptococcus.Front Cell Infect Microbiol. 2014 Aug 4;4:105. doi: 10.3389/fcimb.2014.00105. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25136516 Free PMC article. Review.
-
The two faces of Janus: virulence gene regulation by CovR/S in group A streptococci.Mol Microbiol. 2007 Apr;64(1):34-41. doi: 10.1111/j.1365-2958.2007.05649.x. Mol Microbiol. 2007. PMID: 17376070 Review.
References
-
- Allaw F, Wehbe S, Kanj SS (2024) Necrotizing fasciitis: an update on epidemiology, diagnostic methods, and treatment. Curr Opin Infect Dis 37:105–111 - PubMed
-
- Anand A, Sharma A, Ravins M, Biswas D, Ambalavanan P, Lim KXZ, Tan RYM, Johri AK, Tirosh B, Hanski E (2021) Unfolded protein response inhibitors cure group A streptococcal necrotizing fasciitis by modulating host asparagine. Sci Transl Med 13:eabd7465 - PubMed
-
- Anaya DA, Dellinger EP (2007) Necrotizing soft-tissue infection: diagnosis and management. Clin Infect Dis 44:705–710 - PubMed
-
- Bagcchi S (2023) Surge of invasive group A streptococcus disease. Lancet Infect Dis 23:284 - PubMed
MeSH terms
Substances
Grants and funding
- 1926/24/Israel Science Foundation (ISF)
- 2023YFE0113500/National Key R&D Program from the Ministry of Science and Technology of China
- R01-AI047928/HHS | NIH | NIAID | Division of Microbiology and Infectious Diseases (DMID)
- 82202525/Chinese National Natural Science Foundation
- R01 AI047928/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources

