Integrated bioinformatics analysis reveals that OPRK1 inhibits ferroptosis and induces enzalutamide resistance in prostate cancer
- PMID: 40229787
- PMCID: PMC11998335
- DOI: 10.1186/s40001-025-02484-9
Integrated bioinformatics analysis reveals that OPRK1 inhibits ferroptosis and induces enzalutamide resistance in prostate cancer
Abstract
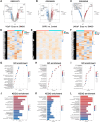
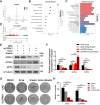
Enzalutamide (Enz) is employed in the management of castration-resistant prostate cancer (CRPC). However, a substantial subset of patients may develop resistance to Enz, thereby reducing its therapeutic effectiveness. The underlying mechanisms contributing to the development of Enz resistance in PCa, whether arising from androgen deprivation or the burden of Enz treatment, remain inadequately understood. OPRK1 plays a key role in Enz resistance through ferroptosis inhibition, which is detected by the analysis of Gene Expression Omnibus (GEO) databases. Silencing OPRK1 via small interfering RNA (siRNA) resulted in the activation of ferroptosis signaling in LNCaP cells. These findings indicate that OPRK1 significantly contributes to Enz resistance in PCa and may serve as a promising therapeutic target for resistant patients.
Keywords: Enzalutamide; Ferroptosis; OPRK1; Prostate cancer.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All procedures performed in this study were in accordance with the ethical standards of the Ethics Committee of The First Hospital of Shanxi Medical University (NO. DWYJ-2023-094) and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
MYO6 contributes to tumor progression and enzalutamide resistance in castration-resistant prostate cancer by activating the focal adhesion signaling pathway.Cell Commun Signal. 2024 Oct 24;22(1):517. doi: 10.1186/s12964-024-01897-z. Cell Commun Signal. 2024. PMID: 39449086 Free PMC article.
-
Bioinformatics analysis reveals that CBX2 promotes enzalutamide resistance in prostate cancer.Eur J Med Res. 2024 Aug 22;29(1):430. doi: 10.1186/s40001-024-02021-0. Eur J Med Res. 2024. PMID: 39175037 Free PMC article.
-
Loss of AR-regulated AFF3 contributes to prostate cancer progression and reduces ferroptosis sensitivity by downregulating ACSL4 based on single-cell sequencing analysis.Apoptosis. 2024 Oct;29(9-10):1679-1695. doi: 10.1007/s10495-024-01941-w. Epub 2024 Mar 13. Apoptosis. 2024. PMID: 38478171
-
The mechanism of histone modifications in regulating enzalutamide sensitivity in advanced prostate cancer.Int J Biol Sci. 2025 Apr 13;21(6):2880-2890. doi: 10.7150/ijbs.109638. eCollection 2025. Int J Biol Sci. 2025. PMID: 40303302 Free PMC article. Review.
-
Prostate cancer: AR aberrations and resistance to abiraterone or enzalutamide.Nat Rev Urol. 2016 Dec;13(12):697-698. doi: 10.1038/nrurol.2016.212. Epub 2016 Nov 2. Nat Rev Urol. 2016. PMID: 27804988 Review. No abstract available.
References
-
- Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12–49. - PubMed
-
- Huang Q, Mitchell C, Theodoulou E, Lee ACK, Brown J. Implementation of fracture risk assessment in men with prostate cancer requiring long-term androgen deprivation therapy: a systematic scoping review using the i-PARIHS implementation framework. J Cancer Surviv. 2024;71(3):209. - PubMed
-
- Akkus E, Arslan Ç, Ürün Y. Advancements in platinum chemotherapy for metastatic castration-resistant prostate cancer: insights and perspectives. Cancer Treat Rev. 2024;130: 102818. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

