Deep targeted sequencing of circulating tumor DNA to inform treatment in patients with metastatic castration-resistant prostate cancer
- PMID: 40229848
- PMCID: PMC11998381
- DOI: 10.1186/s13046-025-03356-0
Deep targeted sequencing of circulating tumor DNA to inform treatment in patients with metastatic castration-resistant prostate cancer
Abstract
Background: Intrinsic and acquired resistance to second-generation anti-androgens pose a significant clinical challenge in the treatment of metastatic castration-resistant prostate cancer (mCRPC). Novel biomarkers to predict treatment response and inform alternative treatment options are urgently needed.
Methods: Deep targeted sequencing, with a prostate cancer-specific gene panel, was performed on circulating tumor DNA (ctDNA) and germline DNA from blood of mCRPC patients recruited in Denmark (n = 53), prior to starting first-line treatment with enzalutamide or abiraterone acetate, and for a subset of patients also at progression (n = 18). Likely clonal hematopoietic variants were filtered out. Genomic findings were correlated to clinical outcomes (PSA progression-free survival (PFS), overall survival (OS)). Intrinsic resistance candidate biomarkers were considered by enrichment analysis of nonresponders vs. responders. Genomic alterations at progression were considered as possible drivers of acquired resistance. Clinical actionability was assessed based on OncoKB and ESCAT.
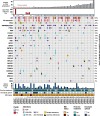
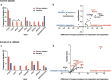
Results: Somatic alterations in PTEN, cell cycle regulators (CCND1, CDKN1B, CDKN2A, and RB1) and chromatin modulators (CHD1, ARID1A) were associated with significantly shorter PFS and OS, also after adjusting for ctDNA% in multivariate Cox regression analysis. The associations with poorer outcomes for alterations in PTEN and chromatin modulators were validated in an external dataset. Patients with primary resistance to enzalutamide/abiraterone had enrichment for BRAF amplification and CHD1 loss, while responders had enrichment for TMPRSS2 fusions. AR resistance mutations emerged in 22% of patients at progression. These were mutually exclusive with other alterations that may confer resistance (i.e., activating CTNNB1 mutations, combined TP53/RB1 loss). Clinically actionable alterations, primarily in homologous recombination repair genes, were found in 54.7% and 49.0% of patients (OncoKB and ESCAT, respectively), with few additional alterations detected at progression. Level I alterations were identified in 41.5% of patients employing OncoKB, however only in 13.2% based on ESCAT.
Conclusions: Our study identifies known and novel prognostic and predictive biomarker candidates in patients with mCRPC undergoing first-line treatment with enzalutamide or abiraterone acetate. It further provides real-world evidence of the significant potential of genomic profiling of ctDNA to inform treatment in this setting. Clinical trials are warranted to advance the implementation of ctDNA-based biomarkers into clinical practice.
Keywords: Biomarker; Circulating cell-free DNA (cfDNA); Circulating tumor DNA (ctDNA); Clinical actionability; Clonal hematopoiesis; Liquid biopsy; Metastatic castration-resistant prostate cancer (mCRPC); Predictive; Prognostic; Resistance.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was approved by the National Committee on Health Research Ethics (#1901101) and the Danish Data Protection Agency (#1–16-02–366-15). All patients provided written informed consent for biobanking. The requirement for patient consent for the specific analyses in this study was waived by the National Committee on Health Research Ethics (#1901101). Consent for publication: Not applicable. Competing interests: Employment or Leadership: M. Borre is chairman for the Danish Prostate Cancer Group and a steering committee member for the Danish Comprehensive Cancer Center. Consultant or Advisory Role: J.B. Jensen has participated on data safety monitoring and/or advisory boards for the following: Roche, Photocure ASA, Olympus, AMBU, Cepheid, and Urotech. H. Grönberg has received consulting fees from Janssen and Astra Zeneca. M. Rusan has participated on advisory boards for Pfizer. Stock Ownership: None declared. Honoraria: J.B. Jensen has received lecturing fees from Olympus and Medac. H. Grönberg has received lecturing fees from Bayer and Astella. M. Borre has received lecturing fees from Astellas, Jansen, Bayer, and MSD. M. Rusan has received lecturing fees from Ipsen and Astra Zeneca, and K.D. Sørensen from Sanofi and “Dagens Medicin.”
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

