Gut microbiota and metabolome signatures in obese and normal-weight patients with colorectal tumors
- PMID: 40230532
- PMCID: PMC11995084
- DOI: 10.1016/j.isci.2025.112221
Gut microbiota and metabolome signatures in obese and normal-weight patients with colorectal tumors
Abstract
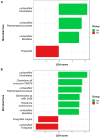
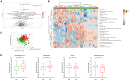
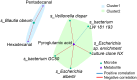
Here, we aim to improve our understanding of various colorectal cancer (CRC) risk factors (obesity, unhealthy diet, and gut microbiota/metabolome alteration), analyzing 120 patients with colon polyps, divided in normal-weight (NW) or overweight/obese (OB). Dietary habits data (validated EPIC questionnaires) revealed a higher consumption of processed meat among OB vs. NW patients. Both mucosa-associated microbiota (MAM) on polyps and lumen-associated microbiota (LAM) analyses uncovered distinct bacterial signatures in the two groups. Importantly, we found an enrichment of the pathogenic species Finegoldia magna in MAM of OB patients, regardless of their polyp stage. We observed distinct mucosal-associated metabolome signatures, with OB patients showing increased pyroglutamic acid and reduced niacin levels, and performed microbiota-metabolome integrated analysis. These findings support a model where different risk factors may contribute to tumorigenesis in OB vs. NW patients, highlighting the potential impact of processed meat consumption and F. magna on CRC development among OB patients.
Keywords: Cancer systems biology; Health sciences; Metabolomics; Microbiome.
© 2025 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Bai H., Xu Z., Li J., Zhang X., Gao K., Fei X., Yang J., Li Q., Qian S., Zhang W., et al. Independent and joint associations of general and abdominal obesity with the risk of conventional adenomas and serrated polyps: A large population-based study in East Asia. Int. J. Cancer. 2023;153:54–63. doi: 10.1002/ijc.34503. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

