Expression of De Novo Open Reading Frames in Natural Populations of Drosophila melanogaster
- PMID: 40231390
- PMCID: PMC12576379
- DOI: 10.1002/jez.b.23297
Expression of De Novo Open Reading Frames in Natural Populations of Drosophila melanogaster
Abstract
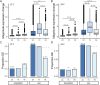
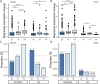
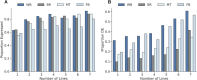
De novo genes, which originate from noncoding DNA, are known to have a high rate of turnover over short evolutionary timescales, such as within a species. Thus, their expression is often lineage- or genetic background-specific. However, little is known about their levels and breadth of expression as populations of a species diverge. In this study, we utilized publicly available RNA-seq data to examine the expression of newly evolved open reading frames (neORFs) in comparison to non- and protein-coding genes in Drosophila melanogaster populations from the derived species range in Europe and the ancestral range in sub-Saharan Africa. Our datasets included two adult tissue types as well as whole bodies at two temperatures for both sexes and three larval/prepupal developmental stages in a single tissue and sex, which allowed us to examine neORF expression and divergence across multiple sample types as well as sex and population. We detected a relatively large proportion (approximately 50%) of annotated neORFs as expressed in the population samples, with neORFs often showing greater expression divergence between populations than non- or protein-coding genes. However, differential expression of neORFs between populations tended to occur in a sample type-specific manner. On the other hand, neORFs displayed less sex-biased expression than the other two gene classes, with the majority of sex-biased neORFs detected in whole bodies, which may be attributable to the presence of the gonads. We also found that neORFs shared among multiple lines in the original set of inbred lines in which they were first detected were more likely to be both expressed and differentially expressed in the new population samples, suggesting that neORFs at a higher frequency (i.e. present in more individuals) within a species are more likely to be functional.
Keywords: de novo genes; gene expression; genome evolution; innovations; novelty; population genetics.
© 2025 The Author(s). Journal of Experimental Zoology Part B: Molecular and Developmental Evolution published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Benjamini, Y. , and Hochberg Y.. 1995. “Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing.” Journal of the Royal Statistical Society Series B: Statistical Methodology 57, no. 1: 289–300. 10.1111/j.2517-6161.1995.tb02031.x. - DOI
-
- Blair, L. , Cridland J., Luo Y., Begun D., and Kopp A.. 2023. “New Insights Into the Dynamics of De Novo Gene Origin.” bioRxiv: 2023‐12. 10.1101/2023.12.08.570739. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

