Cell-intrinsic insulin signaling defects in human iPS cell-derived hepatocytes in type 2 diabetes
- PMID: 40231468
- PMCID: PMC11996863
- DOI: 10.1172/JCI183513
Cell-intrinsic insulin signaling defects in human iPS cell-derived hepatocytes in type 2 diabetes
Abstract
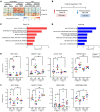
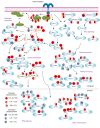
Hepatic insulin resistance is central to type 2 diabetes (T2D) and metabolic syndrome, but defining the molecular basis of this defect in humans is challenging because of limited tissue access. Utilizing inducible pluripotent stem cells differentiated into hepatocytes from control individuals and patients with T2D and liquid chromatography with tandem mass spectrometry-based (LC-MS/MS-based) phosphoproteomics analysis, we identified a large network of cell-intrinsic alterations in signaling in T2D. Over 300 phosphosites showed impaired or reduced insulin signaling, including losses in the classical insulin-stimulated PI3K/AKT cascade and their downstream targets. In addition, we identified over 500 phosphosites of emergent, i.e., new or enhanced, signaling. These occurred on proteins involved in the Rho-GTPase pathway, RNA metabolism, vesicle trafficking, and chromatin modification. Kinome analysis indicated that the impaired phosphorylation sites represented reduced actions of AKT2/3, PKCθ, CHK2, PHKG2, and/or STK32C kinases. By contrast, the emergent phosphorylation sites were predicted to be mediated by increased action of the Rho-associated kinases 1 and 2 (ROCK1/2), mammalian STE20-like protein kinase 4 (MST4), and/or branched-chain α-ketoacid dehydrogenase kinase (BCKDK). Inhibiting ROCK1/2 activity in T2D induced pluripotent stem cell-derived hepatocytes restored some of the alterations in insulin action. Thus, insulin resistance in the liver in T2D did not simply involve a loss of canonical insulin signaling but the also appearance of new phosphorylations representing a change in the balance of multiple kinases. Together, these led to altered insulin action in the liver and identified important targets for the therapy of hepatic insulin resistance.
Keywords: Adult stem cells; Diabetes; Endocrinology; Hepatology; Metabolism.
Conflict of interest statement
Figures





Similar articles
-
Resveratrol Impairs Insulin Signaling in Hepatic Cells via Activation of PKC and PTP1B Pathways.Int J Mol Sci. 2025 Aug 1;26(15):7434. doi: 10.3390/ijms26157434. Int J Mol Sci. 2025. PMID: 40806567 Free PMC article.
-
Modulation of the sodium-chloride cotransporter by insulin in auditory cells: A potential link to diabetes-related hearing complications.J Diabetes Complications. 2025 Sep;39(9):109111. doi: 10.1016/j.jdiacomp.2025.109111. Epub 2025 Jun 22. J Diabetes Complications. 2025. PMID: 40561788
-
Hypoxic extracellular vesicles from hiPSCs protect cardiomyocytes from oxidative damage by transferring antioxidant proteins and enhancing Akt/Erk/NRF2 signaling.Cell Commun Signal. 2024 Jul 9;22(1):356. doi: 10.1186/s12964-024-01722-7. Cell Commun Signal. 2024. PMID: 38982464 Free PMC article.
-
Hybrid closed-loop systems for managing blood glucose levels in type 1 diabetes: a systematic review and economic modelling.Health Technol Assess. 2024 Dec;28(80):1-190. doi: 10.3310/JYPL3536. Health Technol Assess. 2024. PMID: 39673446 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
Cited by
-
Applications and advances of multi-omics technologies in gastrointestinal tumors.Front Med (Lausanne). 2025 Jul 23;12:1630788. doi: 10.3389/fmed.2025.1630788. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40771479 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

