Virus-associated inflammation imprints an inflammatory profile on monocyte-derived macrophages in the human liver
- PMID: 40231469
- PMCID: PMC11996867
- DOI: 10.1172/JCI175241
Virus-associated inflammation imprints an inflammatory profile on monocyte-derived macrophages in the human liver
Abstract
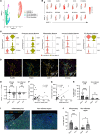
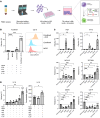
Chronic liver injury triggers the activation and recruitment of immune cells, causing antigen-independent tissue damage and liver disease progression. Tissue inflammation can reshape macrophage composition through monocyte replacement. Replacement of tissue macrophages with monocytes differentiating in an inflammatory environment can potentially imprint a phenotype that switches the liver from an immune-tolerant organ to one predisposed to tissue damage. We longitudinally sampled the liver of patients with chronic hepatitis B who had active liver inflammation and were starting antiviral therapy. Antiviral therapy suppressed viral replication and liver inflammation, which coincided with decreased myeloid activation markers. Single-cell RNA-Seq mapped peripheral inflammatory markers to a monocyte-derived macrophage population, distinct from Kupffer cells, with an inflammatory transcriptional profile. The inflammatory macrophages (iMacs) differentiated from blood monocytes and were unique from macrophage found in healthy or cirrhotic liver. iMacs retained their core transcriptional signature after inflammation resolved, indicating inflammation-mediated remodeling of the macrophage population in the human liver that may affect progressive liver disease and immunotherapy.
Keywords: Hepatitis; Hepatology; Immunology; Infectious disease; Macrophages.
Conflict of interest statement
Figures





References
MeSH terms
LinkOut - more resources
Full Text Sources

