TRIM25-Mediated INSIG1 Ubiquitination Promotes MASH Progression Through Reprogramming Lipid Metabolism
- PMID: 40231613
- PMCID: PMC12140344
- DOI: 10.1002/advs.202414646
TRIM25-Mediated INSIG1 Ubiquitination Promotes MASH Progression Through Reprogramming Lipid Metabolism
Abstract
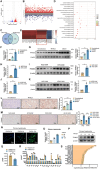
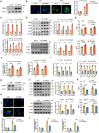
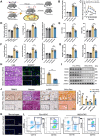
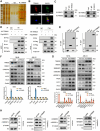
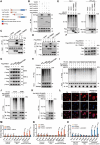
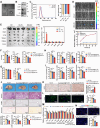
The global incidence of Metabolic dysfunction-associated steatohepatitis (MASH) is increasing, highlighting the urgent need for new treatment strategies. This study aimed to investigate the involvement of tripartite motif-containing 25 (TRIM25) in MASH progression and explore the therapeutic potential of the TRIM25 inhibitor, C27H26N2O4S. Functional studies reveal that TRIM25 promoted lipid accumulation and inflammation by ubiquitinating and degrading insulin-induced gene 1 (INSIG1), thereby enhancing the nuclear translocation of sterol regulatory element-binding protein 2 (SREBP2) and upregulating lipid biosynthesis genes. In vivo experiments using TRIM25 knockout mice demonstrated that TRIM25 deletion ameliorated MASH progression, reduced fibrosis, and decreased inflammatory cell infiltration. It identifies C27H26N2O4S as a specific inhibitor of TRIM25. C27H26N2O4S effectively decreased INSIG1 ubiquitination and attenuated lipid accumulation in the hepatocytes. To enhance the hepatic delivery of C27H26N2O4S, it utilizes exosomes derived from hepatic stellate cells (HSC-EVs). Biodistribution analysis confirmed that the HSC-EVs preferentially accumulated in the liver. In a MASH mouse model, HSC-EV-encapsulated C27H26N2O4S (C27H26N2O4S@HSC-EV) significantly reduced hepatic lipid accumulation and alleviated MASH severity and fibrosis. This study highlights the critical regulatory role of TRIM25 in MASH and presents C27H26N2O4S@HSC-EV as a promising therapeutic approach for MASH treatment.
Keywords: C27H26N2O4S; MASH; TRIM25; exosomes; lipid metabolism; targeted delivery; ubiquitination.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Rinella M. E., Lazarus J. V., Ratziu V., Francque S. M., Sanyal A. J., Kanwal F., Romero D., Abdelmalek M. F., Anstee Q. M., Arab J. P., Arrese M., Bataller R., Beuers U., Boursier J., Bugianesi E., Byrne C. D., Castro Narro G. E., Chowdhury A., Cortez‐Pinto H., Cryer D. R., Cusi K., El‐Kassas M., Klein S., Eskridge W., Fan J., Gawrieh S., Guy C. D., Harrison S. A., Kim S. U.p, Koot B. G., J. Hepatol. 2023, 79, 1542. - PubMed
-
- Rinella M. E., Lazarus J. V., Ratziu V., Francque S. M., Sanyal A. J., Kanwal F., Romero D., Abdelmalek M. F., Anstee Q. M., Arab J. P., Arrese M., Bataller R., Beuers U., Boursier J., Bugianesi E., Byrne C. D., Castro Narro G. E., Chowdhury A., Cortez‐Pinto H., Cryer D. R., Cusi K., El‐Kassas M., Klein S., Eskridge W., Fan J., Gawrieh S., Guy C. D., Harrison S. A., Kim S. U.p, Hepatology. 2023, 78, 1966.
-
- Targher G., Byrne C. D., Tilg H., Gut. 2024, 73, 691. - PubMed
-
- Pericas J. M., Anstee Q. M., Augustin S., Bataller R., Berzigotti A., Ciudin A., Francque S., Abraldes J. G., Hernandez‐Gea V., Pons M., Reiberger T., Rowe I. A., Rydqvist P., Schabel E., Tacke F., Tsochatzis E. A., Genesca J., Nat. Rev. Gastroenterol. Hepatol. 2024, 21, 809. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
