Nanoparticle-based biosensor integrated with multiple cross-displacement amplification for visual and rapid identification of hepatitis B virus and hepatitis C virus
- PMID: 40231683
- PMCID: PMC12054127
- DOI: 10.1128/spectrum.01738-24
Nanoparticle-based biosensor integrated with multiple cross-displacement amplification for visual and rapid identification of hepatitis B virus and hepatitis C virus
Abstract
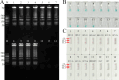
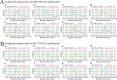
Infection with hepatitis B virus (HBV) and hepatitis C virus (HCV) is a major contributor to liver-related morbidity and mortality worldwide. An accurate and rapid point-of-care (POC) diagnostic approach is the gateway for effective treatment and control of these infections. Here, for the first time, we integrated isothermal multiple cross-displacement amplification (MCDA) with a gold nanoparticle-based lateral flow biosensor (AuNPs-LFB) to successfully develop a novel HBV&HCV-MCDA-AuNPs-LFB assay for simultaneous accurate, sensitive, rapid, inexpensive, and visual identification of HBV and HCV agents. The two unique sets of MCDA degenerate primers were successfully designed targeting the S and 5' untranslated region (5'-UTR) genes from the major HBV genotypes (B, C, D, B/C recombinant, and C/D recombinant) and HCV subtypes in China (1b, 2a, 3a, 3b, and 6a), respectively. The optimal conditions for the MCDA reaction were confirmed to be 64°C for 35 min. The MCDA products were decoded visually using the AuNPs-LFB platform, which was devised for analyzing three targets, including HBV-MCDA, HCV-MCDA amplicons, and a chromatography control. The whole detection procedure, including rapid nucleic acid extraction (~10 min), MCDA reaction (35 min), and AuNPs-LFB interpretation (~2 min), can be completed within 50 min. The HBV&HCV-MCDA-AuNPs-LFB assay can detect the target genes (HBV-S and HCV-5'-UTR) with as low as 10 copies of gene-containing plasmid template per test and does not cross-react with other pathogens. Therefore, our preliminary results indicated that the HBV&HCV-MCDA-AuNPs-LFB assay developed in this study can potentially serve as a useful POC diagnostic tool for the identification of HBV and HCV infections.IMPORTANCEHepatitis B virus (HBV) and hepatitis C virus (HCV) infections have been regarded by the World Health Organization as major threats to human health, especially in low- and middle-income regions. Underdiagnosis of HBV/HCV is a particular challenge for achieving the World Health Organization's goal of eliminating HBV and HCV infections by 2030. Here, for the first time, we integrated isothermal multiple cross-displacement amplification (MCDA) with a gold nanoparticle-based lateral flow biosensor (AuNPs-LFB) to successfully develop a novel HBV&HCV-MCDA-AuNPs-LFB assay for simultaneous accurate, sensitive, rapid, inexpensive, and visual identification and differentiation of HBV and HCV agents.
Keywords: biosensor; hepatitis B virus; hepatitis C virus; lateral flow platform; multiple cross displacement amplification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
One-step, rapid, nanoparticle-based biosensor platform for the simultaneous identification of hepatitis B virus and hepatitis C virus in clinical applications.BMC Microbiol. 2024 Nov 6;24(1):455. doi: 10.1186/s12866-024-03610-z. BMC Microbiol. 2024. PMID: 39506675 Free PMC article.
-
Biosensor-based multiple cross displacement amplification platform for visual and rapid identification of hepatitis C virus.J Med Virol. 2024 Mar;96(3):e29481. doi: 10.1002/jmv.29481. J Med Virol. 2024. PMID: 38425184
-
Rapid, visual, label-based biosensor platform for identification of hepatitis C virus in clinical applications.BMC Microbiol. 2024 Feb 28;24(1):68. doi: 10.1186/s12866-024-03220-9. BMC Microbiol. 2024. PMID: 38413863 Free PMC article.
-
[Markers of hepatitis virus].Rinsho Byori. 2008 Nov;56(11):1014-8. Rinsho Byori. 2008. PMID: 19086457 Review. Japanese.
-
Mini review: current molecular methods for the detection and quantification of hepatitis B virus, hepatitis C virus, and human immunodeficiency virus type 1.Int J Infect Dis. 2014 Aug;25:145-9. doi: 10.1016/j.ijid.2014.04.007. Epub 2014 Jun 10. Int J Infect Dis. 2014. PMID: 24927665 Review.
References
-
- Alberts CJ, Clifford GM, Georges D, Negro F, Lesi OA, Hutin YJ-F, de Martel C. 2022. Worldwide prevalence of hepatitis B virus and hepatitis C virus among patients with cirrhosis at country, region, and global levels: a systematic review. Lancet Gastroenterol Hepatol 7:724–735. doi:10.1016/S2468-1253(22)00050-4 - DOI - PMC - PubMed
-
- Cui F, Blach S, Manzengo Mingiedi C, Gonzalez MA, Sabry Alaama A, Mozalevskis A, Séguy N, Rewari BB, Chan P-L, Le L, Doherty M, Luhmann N, Easterbrook P, Dirac M, de Martel C, Nayagam S, Hallett TB, Vickerman P, Razavi H, Lesi O, Low-beer D. 2023. Global reporting of progress towards elimination of hepatitis B and hepatitis C. Lancet Gastroenterol Hepatol 8:332–342. doi:10.1016/S2468-1253(22)00386-7 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

