Transcription factors DksA and PsrA are synergistic contributors to Legionella pneumophila virulence in Acanthamoeba castellanii protozoa
- PMID: 40231716
- PMCID: PMC12282333
- DOI: 10.1099/mic.0.001551
Transcription factors DksA and PsrA are synergistic contributors to Legionella pneumophila virulence in Acanthamoeba castellanii protozoa
Abstract
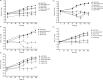
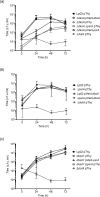
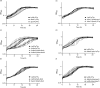
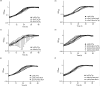
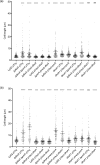
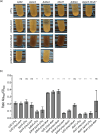
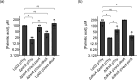
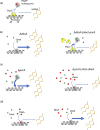
The environmental bacterium Legionella pneumophila, an intracellular parasite of free-living freshwater protozoa as well as an opportunistic human pathogen, has a biphasic lifestyle. The switch from the vegetative replicative form to the environmentally resilient transmissive phase form is governed by a complex stringent response-based regulatory network that includes RNA polymerase co-factor DksA. Here, we report that, through a dysfunctional DksA mutation (DksA1), a synergistic interplay was discovered between DksA and transcription regulator PsrA using the Acanthamoeba castellanii protozoan infection model. Surprisingly, in trans expression of PsrA partially rescued the growth defect of a dksA1 strain. Whilst in trans expression of DksA expectantly could fully rescue the growth defect of the dksA1 strain, it could also surprisingly rescue the growth defect of a ΔpsrA strain. Conversely, the severe intracellular growth defect of a ΔdksA strain could be rescued by in trans expression of DksA and DksA1, but not PsrA. In vitro phenotypic assays show that either DksA or DksA1 was required for extended culturability of bacterial cells, but normal cell morphology and pigmentation required DksA only. Comparative structural modelling predicts that the DksA1 mutation affects the coordination of Mg2+ into the active site of RNAP, compromising transcription efficiency. Taken together, we propose that PsrA transcriptionally assists DksA in the expression of select transmissive phase traits. Additionally, in vitro evidence suggests that the long-chain fatty acid metabolic response is mediated by PsrA together with DksA, inferring a novel regulatory link to the stringent response pathway.
Keywords: Legionella pneumophila; biphasic lifestyle; host-microbe interaction; regulatory network; stringent response; transcription factor.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures




References
-
- Rowbotham TJ. Current views on the relationships between amoebae, legionellae and man. Isr J Med Sci. 1986;22:678–689. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

