Insertion sequence-mediated phage resistance contributes to attenuated colonization of cytolytic Enterococcus faecalis variants in the gut
- PMID: 40231830
- PMCID: PMC12054073
- DOI: 10.1128/spectrum.03303-24
Insertion sequence-mediated phage resistance contributes to attenuated colonization of cytolytic Enterococcus faecalis variants in the gut
Abstract
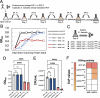
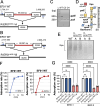
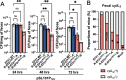
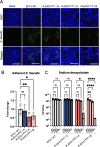
Specific elimination of cytolytic Enterococcus faecalis from the intestinal microbiota by bacteriophages (phages) attenuates ethanol-induced liver disease in pre-clinical studies; however, other clinical phage therapy studies have reported the occurrence of phage-resistant variants. Here, we assessed phage resistance using a cytolytic E. faecalis clinical isolate, EF01. After infecting EF01 with ΦEf2.1 (Myoviridae) or ΦEf2.2 (Podoviridae), four host variants (R-EF01ΦEf2.1-A and R-EF01ΦEf2.1-B from infection with ΦEf2.1, and R-EF01ΦEf2.2-A and R-EF01ΦEf2.2-B from infection with ΦEf2.2) were isolated. Although isolate R-EF01ΦEf2.2 exhibited resistance to both phages, isolate R-EF01ΦEf2.1 demonstrated partial resistance only to ΦEf2.1. Whole-genome sequencing of these four isolates revealed an insertion sequence, IS256, -mediated disruption of xylA in R-EF01ΦEf2.1-A and R-EF01ΦEf2.1-B. In addition, a non-synonymous mutation in epaR, essential for the complete Enterococcus polysaccharide antigen (Epa), was identified in the R-EF01ΦEf2.2-A isolate. Furthermore, R-EF01ΦEf2.2 isolates exhibited IS256-associated chromosomal deletions and lacked galE, a gene involved in Epa biosynthesis. After gavaging mice with EF01 WT, R-EF01ΦEf2.1-A, R-EF01ΦEf2.2-A, and R-EF01ΦEf2.2-B isolates, colonization of R-EF01ΦEf2.2 isolates was significantly attenuated. R-EF01ΦEf2.2 isolates exhibited less resistance to the bile salt sodium deoxycholate and showed reduced adherence to intestinal cell monolayers, suggesting that phage-resistant variants with alterations in bacterial surface molecules, potentially including those involved in Epa biosynthesis, reduced pathogen fitness by attenuating gut colonization. In summary, IS256 is involved in phage resistance of a cytolytic E. faecalis clinical isolate, and certain phage resistance mechanisms could contribute to favorable clinical outcomes by promoting the swift elimination of phage-resistant variants in the treatment of alcohol-associated hepatitis.
Importance: Phage therapy is a promising approach for precise editing of the gut microbiota. Notably, the specific elimination of cytolytic E. faecalis from the intestinal microbiota by phages attenuates ethanol-induced liver disease in pre-clinical studies. Despite the great promise of phage therapy, the occurrence of phage-resistant variants represents a concern for the successful development of phage-based therapies. In this context, we assessed phage resistance using a cytolytic E. faecalis clinical isolate. Isolated phage-resistant variants acquired mutations or deletions of Epa biosynthesis-related genes and exhibited attenuated colonization in the gut. These phage-resistant variants showed less resistance to bile salts and reduced adherence to intestinal cell monolayers. These results suggest that even if phage-resistant variants arise during phage therapy, certain mechanisms of phage resistance may contribute to the rapid elimination of phage-resistant variants promoting favorable clinical outcomes in the treatment of alcohol-associated hepatitis.
Keywords: bacteriophage; cytolysin; fitness cost; gut-liver axis; microbiome editing; phage therapy; trade-off.
Conflict of interest statement
B.S. has been consulting for Ambys Medicines, Ferring Research Institute, Gelesis, HOST Therabiomics, Intercept Pharmaceuticals, Mabwell Therapeutics, Patara Pharmaceuticals, Surrozen, and Takeda. B.S. is the founder of Nterica Bio. UC San Diego has filed several patents with J.F., C.L., and B.S. as inventors related to this work. B.S.'s institution UC San Diego has received research support from Axial Biotherapeutics, BiomX, ChromoLogic, CymaBay Therapeutics, Intercept Pharmaceuticals, NGM Biopharmaceuticals, Prodigy Biotech, and Synlogic Operating Company.
Figures


Similar articles
-
Fully Characterized Effective Bacteriophages Specific against Antibiotic-Resistant Enterococcus faecalis, the Causative Agent of Dental Abscess.Medicina (Kaunas). 2024 Mar 19;60(3):501. doi: 10.3390/medicina60030501. Medicina (Kaunas). 2024. PMID: 38541227 Free PMC article.
-
Bacteriophage Resistance Alters Antibiotic-Mediated Intestinal Expansion of Enterococci.Infect Immun. 2019 May 21;87(6):e00085-19. doi: 10.1128/IAI.00085-19. Print 2019 Jun. Infect Immun. 2019. PMID: 30936157 Free PMC article.
-
Phage Cocktails Constrain the Growth of Enterococcus.mSystems. 2022 Aug 30;7(4):e0001922. doi: 10.1128/msystems.00019-22. Epub 2022 Jun 28. mSystems. 2022. PMID: 35762793 Free PMC article.
-
Evaluation of Phage Therapy in the Context of Enterococcus faecalis and Its Associated Diseases.Viruses. 2019 Apr 20;11(4):366. doi: 10.3390/v11040366. Viruses. 2019. PMID: 31010053 Free PMC article. Review.
-
Phage therapy: A renewed approach against oral diseases caused by Enterococcus faecalis infections.Microb Pathog. 2024 Apr;189:106574. doi: 10.1016/j.micpath.2024.106574. Epub 2024 Feb 13. Microb Pathog. 2024. PMID: 38354990 Review.
Cited by
-
Functional and genomic analysis of Enterococcus phage A155: a potential agent for reducing VRE gut colonization.Virol J. 2025 Jul 30;22(1):263. doi: 10.1186/s12985-025-02849-w. Virol J. 2025. PMID: 40739260 Free PMC article.
-
Phage Therapy: Combating Evolution of Bacterial Resistance to Phages.Viruses. 2025 Aug 8;17(8):1094. doi: 10.3390/v17081094. Viruses. 2025. PMID: 40872808 Free PMC article. Review.
References
-
- Dominguez M, Rincón D, Abraldes JG, Miquel R, Colmenero J, Bellot P, García-Pagán JC, Fernández R, Moreno M, Bañares R, Arroyo V, Caballería J, Ginès P, Bataller R. 2008. A new scoring system for prognostic stratification of patients with alcoholic hepatitis. Am J Gastroenterol 103:2747–2756. doi:10.1111/j.1572-0241.2008.02104.x - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

