Proteomic characterization of Mycobacterium tuberculosis subjected to carbon starvation
- PMID: 40231840
- PMCID: PMC12090744
- DOI: 10.1128/msystems.01530-24
Proteomic characterization of Mycobacterium tuberculosis subjected to carbon starvation
Abstract
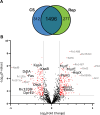
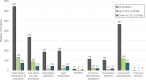
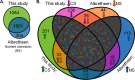
Mycobacterium tuberculosis (Mtb) is the causative agent of tuberculosis (TB), the leading cause of infectious disease-related deaths worldwide. TB infections present on a spectrum from active to latent disease. In the human host, Mtb faces hostile environments, such as nutrient deprivation, hypoxia, and low pH. Under these conditions, Mtb can enter a dormant, but viable, state characterized by a lack of cell replication and increased resistance to antibiotics. Dormant Mtb poses a major challenge to curing infections and eradicating TB globally. We subjected Mtb mc26020 (ΔlysA and ΔpanCD), a double auxotrophic strain, to carbon starvation (CS), a culture condition that induces growth stasis and mimics environmental conditions associated with dormancy in vivo. We provide a detailed analysis of the proteome in CS compared to replicating samples. We observed extensive proteomic reprogramming, with 36% of identified proteins significantly altered in CS. Many enzymes involved in oxidative phosphorylation and lipid metabolism were retained or more abundant in CS. The cell wall biosynthetic machinery was present in CS, although numerous changes in the abundance of peptidoglycan, arabinogalactan, and mycolic acid biosynthetic enzymes likely result in pronounced remodeling of the cell wall. Many clinically approved anti-TB drugs target cell wall biosynthesis, and we found that these enzymes were largely retained in CS. Lastly, we compared our results to those of other dormancy models and propose that CS produces a physiologically distinct state of stasis compared to hypoxia in Mtb.IMPORTANCETuberculosis is a devastating human disease that kills over 1.2 million people a year. This disease is caused by the bacterial pathogen Mycobacterium tuberculosis (Mtb). Mtb excels at surviving in the human host by entering a non-replicating, dormant state. The current work investigated the proteomic changes that Mtb undergoes in response to carbon starvation, a culture condition that models dormancy. The authors found broad effects of carbon starvation on the proteome, with the relative abundance of 37% of proteins significantly altered. Protein changes related to cell wall biosynthesis, metabolism, and drug susceptibility are discussed. Proteins associated with a carbon starvation phenotype are identified, and results are compared to other dormancy models, including hypoxia.
Keywords: Mycobacterium tuberculosis; antibiotic resistance; dormancy; infectious disease; metabolism; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Update of
-
Proteomic characterization of Mycobacterium tuberculosis subjected to carbon starvation.bioRxiv [Preprint]. 2024 Nov 12:2024.11.12.623260. doi: 10.1101/2024.11.12.623260. bioRxiv. 2024. Update in: mSystems. 2025 May 20;10(5):e0153024. doi: 10.1128/msystems.01530-24. PMID: 39605331 Free PMC article. Updated. Preprint.
References
-
- World Health Organization . 2024. Global tuberculosis report 2024. Geneva
-
- Stop TB Partnership . 2022. Global plan to end TB 2023-2030
-
- Grant SS, Kawate T, Nag PP, Silvis MR, Gordon K, Stanley SA, Kazyanskaya E, Nietupski R, Golas A, Fitzgerald M, Cho S, Franzblau SG, Hung DT. 2013. Identification of novel inhibitors of nonreplicating Mycobacterium tuberculosis using a carbon starvation model. ACS Chem Biol 8:2224–2234. doi:10.1021/cb4004817 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
