Recurrent spontaneous miscarriages from sperm after ABVD chemotherapy in a patient with Hodgkin's lymphoma: sperm DNA and methylation profiling
- PMID: 40232270
- PMCID: PMC12422577
- DOI: 10.4103/aja2024107
Recurrent spontaneous miscarriages from sperm after ABVD chemotherapy in a patient with Hodgkin's lymphoma: sperm DNA and methylation profiling
Abstract
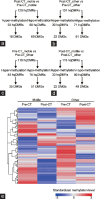
Lymphomas represent one of the most common malignant diseases in young men and an important issue is how treatments will affect their reproductive health. It has been hypothesized that chemotherapies, similarly to environmental chemicals, may alter the spermatogenic epigenome. Here, we report the genomic and epigenomic profiling of the sperm DNA from a 31-year-old Hodgkin lymphoma patient who faced recurrent spontaneous miscarriages in his couple 11-26 months after receiving chemotherapy with adriamycin, bleomycin, vinblastine, and dacarbazine (ABVD). In order to capture the potential deleterious impact of the ABVD treatment on mutational and methylation changes, we compared sperm DNA before and 26 months after chemotherapy with whole-genome sequencing (WGS) and reduced representation bisulfite sequencing (RRBS). The WGS analysis identified 403 variants following ABVD treatment, including 28 linked to genes crucial for embryogenesis. However, none were found in coding regions, indicating no impact of chemotherapy on protein function. The RRBS analysis identified 99 high-quality differentially methylated regions (hqDMRs) for which methylation status changed upon chemotherapy. Those hqDRMs were associated with 87 differentially methylated genes, among which 14 are known to be important or expressed during embryo development. While no variants were detected in coding regions, promoter regions of several genes potentially important for embryo development contained variants or displayed an altered methylated status. These might in turn modify the corresponding gene expression and thus affect their function during key stages of embryogenesis, leading to potential developmental disorders or miscarriages.
Keywords: chemotherapy; embryo loss; methylation; miscarriage; sperm DNA; whole-genome sequencing.
Copyright ©The Author(s)(2025).
Conflict of interest statement
All authors declare no competing interests.
Figures




References
-
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, et al. Cancer incidence and mortality worldwide:sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. - PubMed
-
- National Estimates of Cancer Incidence and Mortality in Metropolitan France between 1990 and 2018 – Malignant Hematological Diseases: A Study Based on Data from the Francim Cancer Registry Network. [[Last accessed on 2024 Dec 25]]. Available from: https://www.santepubliquefrance.fr/docs/estimations-nationales-de-l-inci... .
-
- Shankland KR, Armitage JO, Hancock BW. Non-Hodgkin lymphoma. Lancet. 2012;380:848–57. - PubMed
-
- Ansell SM. Hodgkin lymphoma:diagnosis and treatment. Mayo Clin Proc. 2015;90:1574–83. - PubMed
-
- Fanale MA, Younes A. Monoclonal antibodies in the treatment of non-Hodgkin's lymphoma. Drugs. 2007;67:333–50. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical

