Identifying patients at high risk for antibiotic treatment following hospital admission: a predictive score to improve antimicrobial stewardship measures
- PMID: 40232662
- PMCID: PMC12460503
- DOI: 10.1007/s15010-025-02525-9
Identifying patients at high risk for antibiotic treatment following hospital admission: a predictive score to improve antimicrobial stewardship measures
Abstract
Purpose: Identifying patients for clinical studies evaluating strategies to reduce unnecessary antibiotic usage in hospitals is challenging. This study aimed to develop a predictive score to identify newly hospitalized patients with high likelihood of receiving antibiotics, thus improving patient inclusion in future studies focusing on antimicrobial stewardship (AMS) programs.
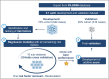
Methods: This retrospective analysis used data from the PILGRIM study (NCT03765528), which included 1,600 patients across ten international sites. Predictive variables for antibiotic treatment during hospitalization were computed, and an additive score model was developed using logistic regression and 10-fold cross-validation. The PILGRIM score was validated in an independent cohort (validation cohort), with performance metrics assessed.
Results: Data from 1,258 patients was included. In the development cohort 52.8% (n = 445) and in the validation cohort 42.4% (n = 134) of patients received antibiotics. Key predictors included hematologic malignancies, immunosuppressive medication, and past hospitalization. The logistic regression model demonstrated an area under the curve of 0.74 in the validation. The final additive score incorporated these predictors plus "planned elective surgery" achieving a specificity of 92%, a positive predictive value of 78%, a sensitivity of 41%, and a negative predictive value (NPV) of 69%in validation set.
Conclusion: The PILGRIM score effectively identifies newly hospitalized patients likely to receive antibiotics, demonstrating high specificity and PPV. Its application can improve future AMS programs and trial recruitment by facilitating targeted inclusion of patients, especially in the hematological and oncological setting. Further -external and prospective- validation is needed to broaden the model's applicability.
Keywords: Antibiotic treatment; Antimicrobial stewardship; Clinical trial; Prediction score.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: MJGTV declares to have received grants or contracts from MSD, Heel, BioNTech, Roche, SD Biosensor, Tillotts. MJGTV received consulting fees from Ferring, Tillotts, Bioaster and payment or honoraria for lectures(presentations, speakers bureaus, manuscript writing or educational events from Akademie für Ärztliche Fort- und Weiterbildung, Akadmie für Infektionsmedizin, Astra Zeneca, bioMerieux, DGI, EUMEDICA, European Society of Neurogastroenterology, Falk Foundation, Ferring, FomF GmbH, Förderkreis Malteser, Frankfurter Bürger Universität, GILEAD, GSK, Helios Kliniken, Hessisches Landessozialgericht, Janssen Cilag GmbH, Jörg Eikerle Beratung, Klinikum Leverkusen, Lahn-Dill Kliniken, Landesärztekammer Hessen, LMU Kliniken, Med. Gesellschaft Bad Homburg, MSD, Pfizer, St. Vincent Hospital, Tillotts. All other authors do not have any conflicts of interest with relevance to this work. Ethics approval and consent to participate: The PILGRIM study was approved by the ethics committees of all participating sites (ID of lead committee in Cologne: UKK 18 - 316) and written informed consent of all participants was obtained prior to any study related measure. The study was conducted in accordance with the Declaration of Helsinki. The study is registered under ClinicalTrials.gov (ID: NCT03765528).
Figures





References
-
- Liss BJ, et al. Intestinal colonisation and blood stream infections due to vancomycin-resistant enterococci (VRE) and extended-spectrum beta-lactamase-producing Enterobacteriaceae (ESBLE) in patients with haematological and oncological malignancies. Infection. 2012;40(6):613–9. - PubMed
-
- Christaki E, Marcou M, Tofarides A. Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J Mol Evol. 2020;88(1):26–40. - PubMed
-
- Harbarth S, et al. Antimicrobial resistance: one world, one fight! Antimicrob Resist Infect Control. 2015;4(1):49.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

