Comparative genomic analysis of paired clinical isolates from a patient with recurrent melioidosis reveals a low within-host mutation rate
- PMID: 40232806
- PMCID: PMC12282283
- DOI: 10.1099/jmm.0.002003
Comparative genomic analysis of paired clinical isolates from a patient with recurrent melioidosis reveals a low within-host mutation rate
Abstract
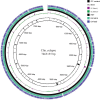
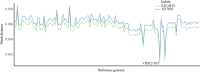
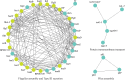
Introduction. Relapse of melioidosis is not uncommon and can occur due to shorter oral antibiotic therapy in the first episode. In such isolates, low mutation rates were identified amongst paired clinical isolates during relapse, but large-scale structural variants were also common.Hypothesis. Using pair-wise comparison, a low number of mutations, especially amongst the virulence and antibiotic resistance genes, may be present amongst the paired isolates obtained during the study period.Aim. A pair of clinical isolates obtained from a patient with recurrent melioidosis during the study period (January 2018 to June 2021) was analysed for identifying the genomic relatedness and DNA changes that may have caused the relapse.Methodology. Using paired-end Illumina sequencing, following appropriate data quality checks, the genomes were assembled using Shovill pipeline, whilst the variants were called using Snippy. Structural variants were detected using TIDDIT, and functional associations were identified using the STRING database searches.Results. One of the isolates (from the second episode) had a highly fragmented genome, but very few structural variants and SNPs were identified. Both the isolates had similar virulence and antibiotic resistance genes; however, owing to the few structural changes, a slightly lower number of virulence genes were observed. Together, they shared 99.8% of the proteomes, and most variants identified spanned either hypothetical proteins or un-annotated regions.Conclusions. Based on comprehensive genome analysis the two strains were genetically similar, with a few structural variants, implying the second episode to be a relapse rather than a re-infection. There was no difference in the antibiotic resistance or virulence genes that may have explained the relapse.
Keywords: Burkholderia pseudomallei; genome sequencing; melioidosis; relapse.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures




Similar articles
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Tracing the environmental source of fatal community-acquired pneumonia and sepsis caused by Burkholderia pseudomallei.Infect Genet Evol. 2025 Sep;133:105788. doi: 10.1016/j.meegid.2025.105788. Epub 2025 Jun 17. Infect Genet Evol. 2025. PMID: 40553791
-
Distinct adaptation and epidemiological success of different genotypes within Salmonella enterica serovar Dublin.Elife. 2025 Jun 25;13:RP102253. doi: 10.7554/eLife.102253. Elife. 2025. PMID: 40560760 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Vitamin D for the treatment of inflammatory bowel disease.Cochrane Database Syst Rev. 2023 Oct 2;10(10):CD011806. doi: 10.1002/14651858.CD011806.pub2. Cochrane Database Syst Rev. 2023. PMID: 37781953 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

