Clinical feature and gene expression analysis in low prostate-specific antigen, high-grade prostate cancer
- PMID: 40233079
- PMCID: PMC11999122
- DOI: 10.1371/journal.pone.0321728
Clinical feature and gene expression analysis in low prostate-specific antigen, high-grade prostate cancer
Abstract
Background: Prostate cancer (PCa) patients with low prostate-specific antigen (PSA) levels can occasionally present high-grade disease. These patients often exhibit resistance to androgen deprivation therapy and have poor outcomes. The mechanisms underlying these observations remain poorly understood. This study aimed to investigate the clinical characteristics and potential gene expression mechanisms in this subgroup.
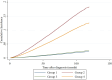
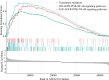
Patients and methods: Clinical data from 365,558 PCa patients were categorized into four groups based on PSA levels and Gleason score (GS): Group 1 (PSA ≤ 2.5 ng/mL, GS < 8), Group 2 (PSA ≤ 2.5 ng/mL, GS ≥ 8), Group 3 (PSA > 2.5 ng/mL, GS < 8), and Group 4 (PSA > 2.5 ng/mL, GS ≥ 8). Clinical characteristics were compared using Kruskal-Wallis H and Pearson's chi-squared tests. Competing-risks regression assessed prostate cancer-specific mortality (PCSM). Gene set enrichment analysis (GSEA) was performed on 219 PCa patients to compare Group A (PSA ≤ 2.5 ng/mL, GS ≥ 8) with Group B (PSA > 2.5 ng/mL, GS ≥ 8).
Results: Group 2 had a significantly higher tumor stage (p < 0.001) and increased hazard ratio for PCSM (p < 0.001). GSEA in Group A identified 156 upregulated gene sets and highlighted several enriched pathways, including the polycomb repressive complex 2, the epidermal growth factor receptor family, retrograde axonal transport, the tumor necrosis factor/nuclear factor-κB pathway, the Rho guanine nucleotide exchange factor/RhoA pathway, and the phosphoinositide 3-kinase signaling pathways (p < 0.05, false discovery rate-adjusted p < 0.25).
Conclusion: PCa patients with low PSA levels and high GS demonstrated an increased risk of PCSM. They were characterized by the aberrant activation of multiple signaling pathways. Targeted therapeutic strategies aimed at these pathways warrant further investigation for their potential to improve outcomes in this aggressive PCa subtype.
Copyright: © 2025 Zhang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Favorable prostate-specific antigen levels correlate with a worse prognosis in high-grade prostate cancer: a population-based analysis.Int J Surg. 2025 Jan 1;111(1):807-817. doi: 10.1097/JS9.0000000000001884. Int J Surg. 2025. PMID: 38935086 Free PMC article.
-
Association of very low prostate-specific antigen levels with increased cancer-specific death in men with high-grade prostate cancer.Cancer. 2016 Jan 1;122(1):78-83. doi: 10.1002/cncr.29691. Epub 2015 Sep 15. Cancer. 2016. PMID: 26371600
-
The U Shape of Prostate-specific Antigen and Prostate Cancer-specific Mortality in High-grade Metastatic Prostate Adenocarcinoma.Eur Urol Focus. 2020 Jan 15;6(1):53-62. doi: 10.1016/j.euf.2018.08.024. Epub 2018 Sep 11. Eur Urol Focus. 2020. PMID: 30217630
-
Global update on defining and treating high-risk localized prostate cancer with leuprorelin: a USA perspective--identifying men at diagnosis who are at high risk of prostate cancer death after surgery or radiation therapy.BJU Int. 2007 Jan;99 Suppl 1:13-6; discussion 17-8. doi: 10.1111/j.1464-410X.2007.06594.x. BJU Int. 2007. PMID: 17229162 Review.
-
Normal Weight, Overweight and Obesity Conditions Associated to Prostate Neoplasm Stages-A Systematic Review and Meta-Analysis.Biomedicines. 2025 May 13;13(5):1182. doi: 10.3390/biomedicines13051182. Biomedicines. 2025. PMID: 40427009 Free PMC article. Review.
References
-
- Schaeffer EM, Srinivas S, Adra N, An Y, Bitting R, Chapin B, et al.. Prostate cancer Guideline (version 4.2024). National Comprehensive Cancer Network. 2024. May 17 [Cited May 19 2024}. Available from: https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf.
-
- Cornford P, van den Bergh RCN, Briers E, Van den Broeck T, Brunckhorst O, Darraugh J, et al.. EAU-EANM-ESTRO-ESUR-ISUP-SIOG guidelines on prostate cancer-2024 update. part I: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2024;86(2):148–63. doi: 10.1016/j.eururo.2024.03.027 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

