Structure of human MUTYH and functional profiling of cancer-associated variants reveal an allosteric network between its [4Fe-4S] cluster cofactor and active site required for DNA repair
- PMID: 40234396
- PMCID: PMC12000561
- DOI: 10.1038/s41467-025-58361-w
Structure of human MUTYH and functional profiling of cancer-associated variants reveal an allosteric network between its [4Fe-4S] cluster cofactor and active site required for DNA repair
Abstract
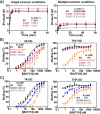
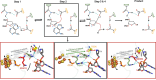
MUTYH is a clinically important DNA glycosylase that thwarts mutations by initiating base-excision repair at 8-oxoguanine (OG):A lesions. The roles for its [4Fe-4S] cofactor in DNA repair remain enigmatic. Functional profiling of cancer-associated variants near the [4Fe-4S] cofactor reveals that most variations abrogate both retention of the cofactor and enzyme activity. Surprisingly, R241Q and N238S retained the metal cluster and bound substrate DNA tightly, but were completely inactive. We determine the crystal structure of human MUTYH bound to a transition state mimic and this shows that Arg241 and Asn238 build an H-bond network connecting the [4Fe-4S] cluster to the catalytic Asp236 that mediates base excision. The structure of the bacterial MutY variant R149Q, along with molecular dynamics simulations of the human enzyme, support a model in which the cofactor functions to position and activate the catalytic Asp. These results suggest that allosteric cross-talk between the DNA binding [4Fe-4S] cofactor and the base excision site of MUTYH regulate its DNA repair function.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Al-Tassan, N. et al. Inherited variants of MYH associated with somatic G: C→ T: a mutations in colorectal tumors. Nat. Genet.30, 227–232 (2002). - PubMed
-
- Messick, T. E. et al. Noncysteinyl coordination to the [4Fe-4S] 2+ cluster of the DNA repair adenine glycosylase MutY introduced via site-directed mutagenesis. Structural characterization of an unusual histidinyl-coordinated cluster. Biochemistry41, 3931–3942 (2002). - PubMed
-
- Porello, S. L., Leyes, A. E. & David, S. S. Single-turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch-containing DNA substrates. Biochemistry37, 14756–14764 (1998). - PubMed
MeSH terms
Substances
Grants and funding
- CHE-2039752/National Science Foundation (NSF)
- GM124169/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 CA067985/CA/NCI NIH HHS/United States
- GM151951/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 GM108583/GM/NIGMS NIH HHS/United States
- S10 OD021527/OD/NIH HHS/United States
- R35 GM151951/GM/NIGMS NIH HHS/United States
- S100D026941/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- P30 GM124165/GM/NIGMS NIH HHS/United States
- GM108583/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- CA069785/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- CHE-1905249/NSF | Directorate for Mathematical & Physical Sciences | Division of Chemistry (CHE)
- CHE-2204229/NSF | Directorate for Mathematical & Physical Sciences | Division of Chemistry (CHE)
- GM124165/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- S100D021527/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- P30 GM124169/GM/NIGMS NIH HHS/United States
- S10 OD026941/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

