Engineering intercellular communication using M13 phagemid and CRISPR-based gene regulation for multicellular computing in Escherichia coli
- PMID: 40234414
- PMCID: PMC12000618
- DOI: 10.1038/s41467-025-58760-z
Engineering intercellular communication using M13 phagemid and CRISPR-based gene regulation for multicellular computing in Escherichia coli
Abstract
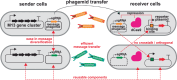
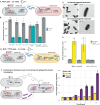
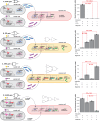
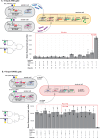
Engineering multicellular consortia, where information processing is distributed across specialized cell types, offers a promising strategy for implementing sophisticated biocomputing systems. However, a major challenge remains in establishing orthogonal intercellular communication, or "wires," within synthetic bacterial consortia. In this study, we address this bottleneck by integrating phagemid-mediated intercellular communication with CRISPR-based gene regulation for multicellular computing in synthetic E. coli consortia. We achieve intercellular communication with high sensitivity by regulating the transfer of single guide RNAs (sgRNAs) encoded on M13 phagemids from sender to receiver cells. Once inside the receiver cells, the transferred sgRNAs mediate gene regulation via CRISPR interference. Leveraging this approach, we successfully constructed one-, two-, and four-input logic gates. Our work expands the toolkit for intercellular communication and paves the way for complex information processing in synthetic microbial consortia, with diverse potential applications, including biocomputing, biosensing, and biomanufacturing.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Phage-mediated intercellular CRISPRi for biocomputation in bacterial consortia.Nucleic Acids Res. 2025 Jan 24;53(3):gkae1256. doi: 10.1093/nar/gkae1256. Nucleic Acids Res. 2025. PMID: 39727169 Free PMC article.
-
Multiplexing cell-cell communication.Mol Syst Biol. 2020 Jul;16(7):e9618. doi: 10.15252/msb.20209618. Mol Syst Biol. 2020. PMID: 32672881 Free PMC article.
-
Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells.Nat Biotechnol. 2015 Sep;33(9):985-989. doi: 10.1038/nbt.3290. Epub 2015 Jun 29. Nat Biotechnol. 2015. PMID: 26121415 Free PMC article.
-
CRISPR Tools for Engineering Prokaryotic Systems: Recent Advances and New Applications.Annu Rev Chem Biomol Eng. 2024 Jul;15(1):389-430. doi: 10.1146/annurev-chembioeng-100522-114706. Epub 2024 Jul 3. Annu Rev Chem Biomol Eng. 2024. PMID: 38598861 Review.
-
Engineering CRISPR guide RNA riboswitches for in vivo applications.Curr Opin Biotechnol. 2019 Feb;55:103-113. doi: 10.1016/j.copbio.2018.08.007. Epub 2018 Sep 25. Curr Opin Biotechnol. 2019. PMID: 30265865 Review.
References
-
- Cubillos-Ruiz, A. et al. Engineering living therapeutics with synthetic biology. Nat. Rev. Drug Discov.20, 941–960 (2021). - PubMed
-
- Wurtzel, E. T. et al. Revolutionizing agriculture with synthetic biology. Nat. Plants5, 1207–1210 (2019). - PubMed
-
- Siuti, P., Yazbek, J. & Lu, T. K. Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol.31, 448–452 (2013). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

