Inhibitory effects of benzyl isothiocyanate on widespread mcr-1-harbouring IncX4 plasmid transfer
- PMID: 40234663
- PMCID: PMC12000558
- DOI: 10.1038/s41598-025-97424-2
Inhibitory effects of benzyl isothiocyanate on widespread mcr-1-harbouring IncX4 plasmid transfer
Abstract
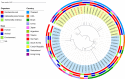
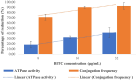
The global dissemination of mobile colistin resistance (mcr) genes represents a significant public health threat due to colistin's critical role in treating multidrug-resistant (MDR) bacterial infections. We identified high rates of carbapenem resistance in Escherichia coli (27.82%) and Klebsiella pneumoniae (57.98%) and colistin resistance in E. coli (7.52%) and K. pneumoniae (19.68%) among MDR clinical isolates in Thailand. We reported sequences of self-transferable IncX4 plasmids (~ 34 kb) that facilitated the spread of the mcr-1.1 gene among six diverse MDR strains, often co-transferring blaCTX-M-55. Additionally, E. coli ST101 was found to co-transfer mcr-1.1, mcr-3.5, blaCTX-M-55, and tet(X4) via three plasmids (~ 34-kb IncX4, ~ 84-kb IncFII, ~ 278-kb IncHI2), resulting in increases in MICs for colistin, ceftriaxone, and tigecycline. Core SNP analysis revealed that closely related IncX4 plasmids harbouring mcr-1 (< 35 SNP differences) were reported from at least 12 countries. We first demonstrated the inhibitory effects of benzyl isothiocyanate (BITC) on the conjugation of mcr-1-bearing IncX4 plasmids to 1.57 ± 1.00% to 48.86 ± 12.31% relative to control (100%), targeting VirB4 and VirB11 proteins, reducing ATPase activity by over 30%. This study highlights the widespread mcr-1-harbouring IncX4 plasmids and proposes BITC as a potential inhibitor to control the dissemination of colistin resistance.
Keywords: Escherichia coli; Klebsiella pneumoniae; Benzyl isothiocyanate; Colistin; Plasmids; Thailand.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics approval: The study was conducted by the Biosafety guidelines and approved by the Mahidol University-Institutional Biosafety Committee (MU-IBC), Mahidol University (Nakhon Pathom, Thailand) [MU 2023-009]. This study of anonymised clinical isolates did not include interaction or intervention with human subjects or access to identifiable private information. The Mahidol University Central Institutional Review Board (MU-CIRB), Mahidol University (Nakhon Pathom, Thailand) waived the requirement for IRB review. Informed consent: For this retrospective study of anonymous clinical isolates, the requirement for informed consent from patients was waived by the Mahidol University Central Institutional Review Board (MU-CIRB), Mahidol University (Nakhon Pathom, Thailand).
Figures




References
-
- Liu, J. H. et al. Plasmid-mediated colistin-resistance genes: mcr. Trends Microbiol32, 365–378. 10.1016/j.tim.2023.10.006 (2024). - PubMed
-
- Schwarz, S. & Johnson, A. P. Transferable resistance to colistin: A new but old threat. J Antimicrob Chemother71, 2066–2070. 10.1093/jac/dkw274 (2016). - PubMed
-
- Liu, Y. Y. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect Dis16, 161–168. 10.1016/S1473-3099(15)00424-7 (2016). - PubMed
-
- Haenni, M. et al. Co-occurrence of extended spectrum beta lactamase and MCR-1 encoding genes on plasmids. Lancet Infect Dis16, 281–282. 10.1016/S1473-3099(16)00007-4 (2016). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

