Sex-Biased Admixture Followed by Isolation and Adaptive Evolution Shaped the Genomic and Blood Pressure Diversity of the LopNur People
- PMID: 40235149
- PMCID: PMC12034462
- DOI: 10.1093/molbev/msaf091
Sex-Biased Admixture Followed by Isolation and Adaptive Evolution Shaped the Genomic and Blood Pressure Diversity of the LopNur People
Abstract
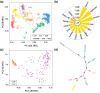
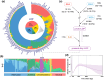
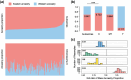
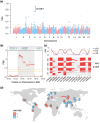
The LopNur people are an ethnic group living on the edge of the Taklamakan Desert, and they are believed to demonstrate a unique genetic makeup due to their isolation and limited contact with neighboring populations. However, a lack of genetic studies on the LopNur people has resulted in limited knowledge about their ancestral origins and demographic history. Here, we conducted the first whole-genome sequencing study of 164 LopNur individuals (LOP) to gain insight into their genetic history and adaptive evolution in an isolated desert area. Our analysis revealed that the present-day LOP have experienced a complex history of admixture followed by long-term isolation, with their ancestry derived from East Asia (∼41.46%), West Eurasia (∼26.43%), Siberia (∼24.27%), and South Asia (∼7.82%). Notably, a remarkable sex-biased admixture occurred between Western males and Eastern females. In addition to complex admixture followed by long-term geographic isolation and further recent migrations, adaptive evolution jointly formed the gene pool and phenotypic diversity of the present-day LOP. Intriguingly, our analysis suggests that the USP35-GAB2 region may be correlated with blood pressure in LOP, based on a joint analysis of genomics and blood pressure data. Moreover, we identified two variants, rs7387065, and rs2229437, located on CSMD1 and PRCP, respectively. These variants exhibited frequency differences between Asian and European populations and were reported to be associated with antihypertensive drug absorption. Our results provide new insight into the complex history of the LOP, an admixed and isolated ethnic group residing at the crossroads of East and West, a case with ancient admixture, long-term isolation, adaptive evolution, and sex-biased gene flow.
Keywords: LopNur people; genetic admixture; hypertension; isolation; local adaptation; precision medicine.
© The Author(s) 2025. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest: The authors declare that they have no competing interests.
Figures

References
-
- Abdurehim E. The LopNor dialect of Uyghur. Helsinki: University of Helsinki Printing House (Unigrafia); 2014.
-
- Ametjan A, Gan GQ. The research review of LopNur people since the reform and opening up. J Kashgar Teachers Coll. 2014:35(1). 10.13933/j.cnki.j.kashgar.teach.coll.2014.01.024. - DOI
-
- Aragam KG, Jiang T, Goel A, Kanoni S, Wolford BN, Atri DS, Weeks EM, Wang M, Hindy G, Zhou W, et al. Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Nat Genet. 2022:54(12):1803–1815. 10.1038/s41588-022-01233-6. - DOI - PMC - PubMed
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

