This is a preprint.
De novo variants in KDM2A cause a syndromic neurodevelopmental disorder
- PMID: 40236430
- PMCID: PMC11998838
- DOI: 10.1101/2025.03.31.25324695
De novo variants in KDM2A cause a syndromic neurodevelopmental disorder
Abstract
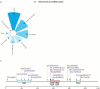
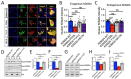
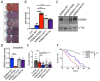
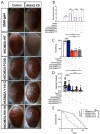
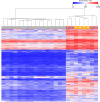
Germline variants that disrupt components of the epigenetic machinery cause syndromic neurodevelopmental disorders. Using exome and genome sequencing, we identified de novo variants in KDM2A, a lysine demethylase crucial for embryonic development, in 18 individuals with developmental delays and/or intellectual disabilities. The severity ranged from learning disabilities to severe intellectual disability. Other core symptoms included feeding difficulties, growth issues such as intrauterine growth restriction, short stature and microcephaly as well as recurrent facial features like epicanthic folds, upslanted palpebral fissures, thin lips, and low-set ears. Expression of human disease-causing KDM2A variants in a Drosophila melanogaster model led to neural degeneration, motor defects, and reduced lifespan. Interestingly, pathogenic variants in KDM2A affected physiological attributes including subcellular distribution, expression and stability in human cells. Genetic epistasis experiments indicated that KDM2A variants likely exert their effects through a potential gain-of-function mechanism, as eliminating endogenous KDM2A in Drosophila did not produce noticeable neurodevelopmental phenotypes. Data from Enzymatic-Methylation sequencing supports the suggested gene-disease association by showing an aberrant methylome profiles in affected individuals' peripheral blood. Combining our genetic, phenotypic and functional findings, we establish de novo variants in KDM2A as causative for a syndromic neurodevelopmental disorder.
Conflict of interest statement
Declaration of interests DAC, LMD and SVM are employees of and may own stock in GeneDx, LLC. The other authors declare no competing interests.
Figures

References
-
- Levy M.A., McConkey H., Kerkhof J., Barat-Houari M., Bargiacchi S., Biamino E., Bralo M.P., Cappuccio G., Ciolfi A., Clarke A., et al. (2022). Novel diagnostic DNA methylation episignatures expand and refine the epigenetic landscapes of Mendelian disorders. Human Genetics and Genomics Advances 3, 100075. 10.1016/j.xhgg.2021.100075. - DOI - PMC - PubMed
-
- Faundes V., Newman W.G., Bernardini L., Canham N., Clayton-Smith J., Dallapiccola B., Davies S.J., Demos M.K., Goldman A., Gill H., et al. (2018). Histone Lysine Methylases and Demethylases in the Landscape of Human Developmental Disorders. The American Journal of Human Genetics 102, 175–187. 10.1016/j.ajhg.2017.11.013. - DOI - PMC - PubMed
-
- van Jaarsveld R.H., Reilly J., Cornips M.-C., Hadders M.A., Agolini E., Ahimaz P., Anyane-Yeboa K., Bellanger S.A., van Binsbergen E., van den Boogaard M.-J., et al. (2022). Delineation of a KDM2B-related neurodevelopmental disorder and its associated DNA methylation signature. Genetics in Medicine, S109836002200942X. 10.1016/j.gim.2022.09.006. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
