This is a preprint.
Inclusion bias affects common variant discovery and replication in a health-system linked biobank
- PMID: 40236437
- PMCID: PMC11998835
- DOI: 10.1101/2025.04.04.25325131
Inclusion bias affects common variant discovery and replication in a health-system linked biobank
Abstract
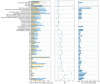
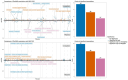
Electronic Health Records (EHR) -linked biobanks have emerged as promising tools for precision medicine, enabling the integration of clinical and molecular data for individual risk assessment. Association studies performed in biobank studies can connect common genetic variation to clinical phenotypes, such as through the use of polygenic scores (PGS), which are starting to have utility in aiding clinician decision making. However, while biobanks aggregate large amounts of data effectively for such studies, most employ various opt-in consent protocols, and, as a result, are expected to be subject to participation and recruitment biases. The extent to which biases affect genetic analyses in biobanks remains unstudied. In this study, we quantify bias and evaluate its impact on genetic analyses, using the UCLA ATLAS Community Health Initiative as a case study. Our analyses reveal that a wide array of factors, particularly socio-demographic characteristics and healthcare utilization patterns, influence participation, effectively differentiating biobank participants from the broader patient population (AUROC = 0.85, AUPRC = 0.82). Through weighting the sample using inverse probability weights derived from probabilities of enrollment, we replicated 54% more known GWAS variants than models that did not take bias into account (e.g. associations between variants in the PPARG gene and type 2 diabetes). We further show that PGS-Phenome wide associations are affected by the weighting scheme, and suggest associations corroborated by weighted analyses to be more robust. Our results highlight that genetic analyses within biobanks should account for inclusion biases, and suggest inverse probability weighting as a potential approach.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
