Fully automatic HER2 tissue segmentation for interpretable HER2 scoring
- PMID: 40236564
- PMCID: PMC11999220
- DOI: 10.1016/j.jpi.2025.100435
Fully automatic HER2 tissue segmentation for interpretable HER2 scoring
Abstract
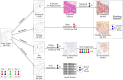
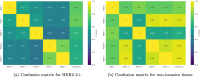
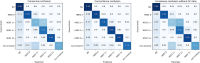
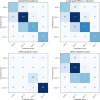
Breast cancer is the most common cancer in women, with HER2 (human epidermal growth factor receptor 2) overexpression playing a critical role in regulating cell growth and division. HER2 status, assessed according to established scoring guidelines, offers important information for treatment selection. However, the complexity of the task leads to variability in human rater assessments. In this work, we propose a fully automated, interpretable HER2 scoring pipeline based on pixel-level semantic segmentations, designed to align with clinical guidelines. Using polygon annotations, our method balances annotation effort with the ability to capture fine-grained details and larger structures, such as non-invasive tumor tissue. To enhance HER2 segmentation, we propose the use of a Wasserstein Dice loss to model class relationships, ensuring robust segmentation and HER2 scoring performance. Additionally, based on observations of pathologists' behavior in clinical practice, we propose a calibration step to the scoring rules, which positively impacts the accuracy and consistency of automated HER2 scoring. Our approach achieves an F1 score of 0.832 on HER2 scoring, demonstrating its effectiveness. This work establishes a potent segmentation pipeline that can be further leveraged to analyze HER2 expression in breast cancer tissue.
Keywords: Deep learning; HER2; HER2 scoring; Histopathology; Semantic segmentation.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Figures




Similar articles
-
AutoIHC-Analyzer: computer-assisted microscopy for automated membrane extraction/scoring in HER2 molecular markers.J Microsc. 2021 Jan;281(1):87-96. doi: 10.1111/jmi.12955. Epub 2020 Aug 27. J Microsc. 2021. PMID: 32803890
-
Interpretable HER2 scoring by evaluating clinical guidelines through a weakly supervised, constrained deep learning approach.Comput Med Imaging Graph. 2023 Sep;108:102261. doi: 10.1016/j.compmedimag.2023.102261. Epub 2023 Jun 15. Comput Med Imaging Graph. 2023. PMID: 37356357
-
Transfer learning drives automatic HER2 scoring on HE-stained WSIs for breast cancer: a multi-cohort study.Breast Cancer Res. 2025 Apr 23;27(1):62. doi: 10.1186/s13058-025-02008-7. Breast Cancer Res. 2025. PMID: 40269991 Free PMC article.
-
Fully Automated Artificial Intelligence Solution for Human Epidermal Growth Factor Receptor 2 Immunohistochemistry Scoring in Breast Cancer: A Multireader Study.JCO Precis Oncol. 2024 Oct;8:e2400353. doi: 10.1200/PO.24.00353. Epub 2024 Oct 11. JCO Precis Oncol. 2024. PMID: 39393036 Free PMC article.
-
Independent Validation of a HER2-Low Focused Immunohistochemistry Scoring System for Enhanced Pathologist Precision and Consistency.Mod Pathol. 2025 Apr;38(4):100693. doi: 10.1016/j.modpat.2024.100693. Epub 2024 Dec 24. Mod Pathol. 2025. PMID: 39724961
References
-
- Ferlay J., Colombet M., Soerjomataram I., et al. Cancer statistics for the year 2020: an overview. Int J Cancer. 2021;149(4):778–789. - PubMed
-
- Loibl S., Gianni L. HER2-positive breast cancer. Lancet. 2017;389(10087):2415–2429. - PubMed
-
- Wolff A.C., Hammond M.E.H., Allison K.H., et al. Human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of american pathologists clinical practice guideline focused update. Arch Pathol Lab Med. 2018;142(11):1364–1382. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

