Biochemical and Biophysical Divergences between Two Escherichia coli l-Asparaginase II Variants: Potential for Using EcA2-K12 as a Biosimilar
- PMID: 40237204
- PMCID: PMC12273708
- DOI: 10.1021/acs.biochem.4c00663
Biochemical and Biophysical Divergences between Two Escherichia coli l-Asparaginase II Variants: Potential for Using EcA2-K12 as a Biosimilar
Abstract
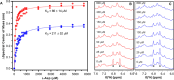
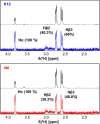
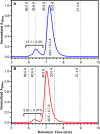
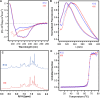
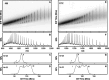
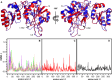
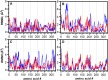
Escherichia coli l-asparaginase II (EcA2) is essential for treating Acute Lymphoblastic Leukemia, the most common childhood cancer. This enzyme catalyzes the hydrolysis of l-asparagine or l-glutamine to ammonia and l-aspartate or l-glutamate. The first FDA-approved EcA2 biopharmaceutical, Elspar, was introduced in 1978, followed by other biosimilars. Despite stringent approval criteria, variations in plasmatic activity and therapeutic efficacy persist across different EcA2 preparations, often leading to substandard product notifications. Many studies focus on the EcA2 from the E. coli K12 strain (EcA2-K12), which differs by four amino acids from reference biopharmaceuticals, including Elspar (EcA2-4M). Here, we show that EcA2-4 M has over twice the specific activity on both the hydrolysis of l-asparagine and on human lymphoblast cells compared to EcA2-K12. EcA2-K12 demonstrates 4-fold greater specificity for l-asparagine over l-glutamine, considering their kcat, but similar KM toward each amino acid. Interestingly, EcA2-K12 has 3-fold lower affinity for l-aspartate, linked to reduced stabilization of its N-terminal active site loop. Although both variants exhibit indistinguishable thermostability, EcA-K12 shows a higher tendency to oligomerize. We solved the 3D structures of both variants by X-ray crystallography, and normal-mode analysis revealed wider conformational changes in EcAK12's active site. Our data indicate that EcA2-K12 has lower activity due to the higher conformational dynamics of the N-terminal active site loop. Nevertheless, EcA2-K12 is a beneficial alternative or complement to existing therapeutic schemes with EcA2-4M, due to its higher specificity to l-asparagine, which is of fundamental importance since activity on l-glutamine is associated with harmful side effects.
Figures




Similar articles
-
Anti-vascular endothelial growth factor biosimilars for neovascular age-related macular degeneration.Cochrane Database Syst Rev. 2024 Jun 3;6(6):CD015804. doi: 10.1002/14651858.CD015804.pub2. Cochrane Database Syst Rev. 2024. PMID: 38829176 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Hyperthermophilic L-Asparaginase from Thermococcus sibiricus and Its Double Mutant with Increased Activity: Insights into Substrate Specificity and Structure.Int J Mol Sci. 2025 Jun 6;26(12):5437. doi: 10.3390/ijms26125437. Int J Mol Sci. 2025. PMID: 40564901 Free PMC article.
-
Systemic Inflammatory Response Syndrome.2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31613449 Free Books & Documents.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
-
- World Health Organization . Model List of Essential Medicines for Children9th List; World Health Organization: Geneva, 2023.
-
- Allemani C., Weir H. K., Carreira H., Harewood R., Spika D., Wang X.-S., Bannon F., Ahn J. V., Johnson C. J., Bonaventure A., Marcos-Gragera R., Stiller C., Azevedo e Silva G., Chen W.-Q., Ogunbiyi O. J., Rachet B., Soeberg M. J., You H., Matsuda T., Bielska-Lasota M., Storm H., Tucker T. C., Coleman M. P.. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25 676 887 patients from 279 population-based registries in 67 countries (CONCORD-2) Lancet. 2015;385:977–1010. doi: 10.1016/S0140-6736(14)62038-9. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

