AUP1 and UBE2G2 complex targets STING signaling and regulates virus-induced innate immunity
- PMID: 40237449
- PMCID: PMC12077101
- DOI: 10.1128/mbio.00602-25
AUP1 and UBE2G2 complex targets STING signaling and regulates virus-induced innate immunity
Abstract
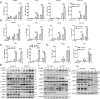
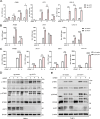
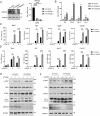
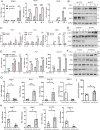
Stimulator of interferon genes (STING) is an endoplasmic reticulum (ER) signaling adaptor that is essential for the host immune response triggered by DNA pathogens. Precise regulation of STING is crucial for maintaining a balanced immune response and preventing harmful autoinflammation. Activation of STING requires its translocation from the ER to the Golgi apparatus. However, the mechanisms that maintain STING in its resting state remain largely unclear. Here, we find that deficiency of the ancient ubiquitous protein 1 (AUP1) causes spontaneous activation of STING and enhances the expression of type I interferons (IFNs) under resting conditions. Furthermore, deficiency of UBE2G2, a cofactor of AUP1, also promotes the abnormal activation of STING. AUP1 deficiency significantly enhances STING signaling induced by DNA virus, and AUP1 deficiency exhibits increased resistance to DNA virus infection in vitro and in vivo. Mechanistically, AUP1 may form a complex with UBE2G2 to interact with STING, preventing its exit from the ER membrane. Notably, infection with the RNA virus vesicular stomatitis virus (VSV) promotes the accumulation of lipid droplets (LDs) and AUP1 proteins. Additionally, AUP1 deficiency markedly inhibits the replication of VSV because AUP1 deficiency reduces lipid accumulation and alters the expression of lipid metabolism genes, such as carnitine palmitoyltransferase 1A (CPT1A), monoglyceride lipase (MGLL), and sterol regulatory element-binding transcription factor 1 (SREBF1). This study uncovers the essential roles of AUP1 in the STING signaling pathway and lipid metabolism pathway, highlighting its dual role in regulating virus replication.IMPORTANCEThe stimulator of interferon genes (STING) signaling cascade plays an essential role in coordinating innate immunity against DNA pathogens and autoimmunity. Precise regulation of the innate immune response is essential for maintaining homeostasis. In this study, we demonstrate that ancient ubiquitous protein 1 (AUP1) and UBE2G2 act as negative regulators of the innate immune response by targeting STING. Notably, AUP1 interacts with STING to retain STING in the endoplasmic reticulum (ER), preventing STING translocation and thereby limiting STING signaling in the resting state. In addition, deficiency of AUP1 markedly inhibits the replication of DNA virus and RNA virus. Our findings provide new insights into the regulation of STING signaling and confirm AUP1 has a dual role in regulating virus replication.
Keywords: AUP1; STING; UBE2G2; innate immunity; virus infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Jeremiah N, Neven B, Gentili M, Callebaut I, Maschalidi S, Stolzenberg M-C, Goudin N, Frémond M-L, Nitschke P, Molina TJ, Blanche S, Picard C, Rice GI, Crow YJ, Manel N, Fischer A, Bader-Meunier B, Rieux-Laucat F. 2014. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J Clin Invest 124:5516–5520. doi: 10.1172/JCI79100 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
