Peculiar morphology of Asgard archaeal cells close to the prokaryote-eukaryote boundary
- PMID: 40237460
- PMCID: PMC12077186
- DOI: 10.1128/mbio.00327-25
Peculiar morphology of Asgard archaeal cells close to the prokaryote-eukaryote boundary
Abstract
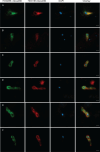
The emergence of complex eukaryotic cells from prokaryotic ancestors is a major enigma in the history of life. Current data suggest that Asgard archaea are the closest prokaryotic relatives of eukaryotes and have a genetic potential for cellular complexity, suggesting their key role in the evolution of eukaryotes. The Asgard archaeal order Hodarchaeales was recently proposed as the sister lineage of eukaryotes with a unique set of eukaryotic signature proteins. However, there is no microscopic evidence to show the cellular structure of these closest known archaeal relatives of eukaryotes. Here, we retrieved Hodarchaeales-affiliated full-length 16S rRNA sequences from marine sediments (Aarhus Bay, Denmark), representing 0.1% of the relative rRNA read abundance in sequence libraries, and designed new oligonucleotide probes specifically targeting their cells. We then employed catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) and imaged the labeled cells by super-resolution microscopy with appropriate controls. Hodarchaeales-affiliated cells were characterized by an elongated cell body connected to a rounded expansion at one pole with a confined central DNA localization. They were conspicuously large, with an average length of 3 µm, ranging from 1.5 to 5.2 µm. The average width was 1.1-0.8 µm in the round expansion and the cell body, respectively. The remarkable size and morphology of the detected Hodarchaeales-related cells suggest a potential for complex eukaryote-like cellular architecture in these as-yet-uncultivated Asgard archaea, which could represent a key transitional stage in eukaryogenesis.IMPORTANCEAsgard archaea played a pivotal role in the evolution of complex cellular life, as recent data revealed their close relationship to eukaryotes in the Tree of Life and a genetic repertoire suggesting intricate cellular organization. This suggests that the key elements of eukaryotic cellular complexity originated in Asgard archaea. However, visual evidence to show their cellular structure and morphological diversity remains scarce, leaving an open question of how the genetic potential for cellular complexity translates into phenotype. In this study, we report the remarkable size and unusual shape of yet-uncultivated Asgard archaeal cells, which are close to the prokaryote-eukaryote boundary. Our findings suggest a previously unknown complex cellular structure in the closest archaeal relatives of eukaryotes and could contribute to our understanding of the evolution of complex life forms.
Keywords: Asgard archaea; Hodarchaeales; cellular complexity; eukaryogenesis; fluorescence microscopy; in situ hybridization; marine sediments.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, Stott MB, Nunoura T, Banfield JF, Schramm A, Baker BJ, Spang A, Ettema TJG. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541:353–358. doi:10.1038/nature21031 - DOI - PubMed
-
- Eme L, Tamarit D, Caceres EF, Stairs CW, De Anda V, Schön ME, Seitz KW, Dombrowski N, Lewis WH, Homa F, Saw JH, Lombard J, Nunoura T, Li W-J, Hua Z-S, Chen L-X, Banfield JF, John ES, Reysenbach A-L, Stott MB, Schramm A, Kjeldsen KU, Teske AP, Baker BJ, Ettema TJG. 2023. Inference and reconstruction of the heimdallarchaeial ancestry of eukaryotes. Nature 618:992–999. doi:10.1038/s41586-023-06186-2 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

