Mass cytometry analysis reveals a cross-tissue immune landscape in Actinobacillus pleuropneumoniae-induced pneumonia
- PMID: 40237529
- PMCID: PMC12131827
- DOI: 10.1128/spectrum.02665-24
Mass cytometry analysis reveals a cross-tissue immune landscape in Actinobacillus pleuropneumoniae-induced pneumonia
Abstract
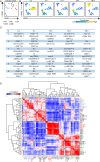
Porcine contagious pleuropneumonia caused by Actinobacillus pleuropneumoniae (APP) is a fatal respiratory disease that threatens the worldwide farming industry's health. The immune responses of extrapulmonary tissues play an important role in developing porcine contagious pleuropneumonia; however, the immune responses of extrapulmonary tissues induced by APP are rarely uncovered. Here, we used high-dimensional mass cytometry to investigate the immune cell response in the spleen and peripheral blood during APP infection in mice. We found that the immune response triggered by APP was highly tissue-specific. Numerous infection time- or tissue-specific immune cell clusters, including previously unrecognized ones, were also identified in the spleen and peripheral blood. Integrative analysis of splenic lymphoid and myeloid cell clusters maps the dynamic immune response cellular network during APP infection. Surprisingly, during the early stages of APP infection, the majority of the top 6 cell clusters contributing to the infection time-specificity in the spleen were adaptive immune cell clusters rather than innate immune cell clusters, among which CD24hiMHCII+CD8+TEM cells exhibited a stronger expression of IFN-γ, IL-17A, and IL-10 compared to the CD24lo compartment. In peripheral blood, there was unprecedented heterogeneity in the immune cell composition. Also, peripheral immune cell clusters closely related to the severity of APP infection were identified. In summary, our data provide a systemic and comprehensive overview of the immune responses to APP infection in the spleen and peripheral blood. This provides a foundation for understanding the immune pathogenesis of APP and identifying potential diagnostic biomarkers and therapeutic targets.
Importance: This study explored the cross-tissue immune dynamic landscape in the APP-induced pneumonia model by utilizing high-dimensional mass cytometry. We discovered that APP-induced immune responses are tissue-specific. Key infection-specific clusters in the spleen and peripheral blood were identified, some of which were previously unrecognized. Meanwhile, the specific functions of APP infection-related immune subsets were explored. The research systematically outlined an overview of immune responses in these tissues, deepening the understanding of APP pathogenesis and laying the foundation for the search for diagnostic and therapeutic targets.
Keywords: Actinobacillus pleuropneumoniae; immune cell clusters; immune response; mass cytometry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Li N, Zhu J, Chen P, Bao C, Wang J, Abdelaal T, Chen D, Zhu S, Wang W, Mao J, Scicluna BP, Koning F, Li F, Lei L. 2023. High-dimensional analysis reveals an immune atlas and novel neutrophil clusters in the lungs of model animals with Actinobacillus pleuropneumoniae-induced pneumonia. Vet Res 54:76. doi: 10.1186/s13567-023-01207-4 - DOI - PMC - PubMed
-
- Nerland EM, LeBlanc JM, Fedwick JP, Morck DW, Merrill JK, Dick P, Paradis MA, Buret AG. 2005. Effects of oral administration of tilmicosin on pulmonary inflammation in piglets experimentally infected with Actinobacillus pleuropneumoniae. Am J Vet Res 66:100–107. doi: 10.2460/ajvr.2005.66.100 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

