FOXR2 activation is not exclusive of CNS neuroblastoma
- PMID: 40237561
- PMCID: PMC12417820
- DOI: 10.1093/neuonc/noaf076
FOXR2 activation is not exclusive of CNS neuroblastoma
Abstract
Background: FOXR2 activation is regarded as pathognomonic for CNS neuroblastoma (NB). However, a comprehensive understanding of the landscape for CNS tumors exhibiting FOXR2 activation is lacking.
Methods: Histopathologic, molecular, imaging, and clinical data of 42 CNS tumors with FOXR2 overexpression identified through screening institutional datasets and published institutional cases were analyzed.
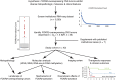
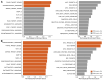
Results: Among the 42 tumors, 21 (50.0%) were high-grade gliomas (HGGs), and 18 (42.9%) were embryonal tumors. The HGGs included ten H3 K27M-mutant diffuse midline gliomas (DMGs) and eight radiation-associated tumors. The embryonal tumors included 11 CNS NBs and six pineoblastomas (PBs). FOXR2 expression was similar between CNS NB and other tumor types (P = 0.82). HGGs with FOXR2 overexpression, unlike NBs and PBs, displayed diverse concomitant genetic alterations. The most common mechanisms of FOXR2 activation involved structural alterations causing promoter donation and enhancer hijacking from active genes essential for brain development, followed by alternative promoter activation or truncated LINE-1 retrotransposition. The preferential activation mechanism varied by tumor type. All but two aberrant FOXR2 transcripts incorporated non-canonical, non-coding exons. Gene set enrichment analysis demonstrated shared downstream effects of FOXR2 activation at the epigenome and transcriptome levels across tumor types. DMGs and PBs with FOXR2 overexpression were aggressive, with 0% 2-year overall survival, whereas CNS NBs responded well to combined chemotherapy and radiation.
Conclusions: CNS tumors with FOXR2 overexpression manifest significant histological, molecular, imaging, and clinical diversity. While HGGs and PBs with FOXR2 overexpression demonstrated inferior prognosis, CNS NBs showed favorable outcomes. Integrating histologic and molecular diagnostic approaches is imperative for accurate prognostication and optimal therapeutic decision-making.
Keywords: CNS tumors with FOXR2 overexpression; LINE-1 retrotransposition; alternative promoter; patient outcomes; promoter donation and enhancer hijacking.
© The Author(s) 2025. Published by Oxford University Press on behalf of the Society for Neuro-Oncology.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Gharbaran R. Insights into the molecular roles of FOXR2 in the pathology of primary pediatric brain tumors. Crit Rev Oncol Hematol. 2023;192:104188. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

