Transcriptome Meta-Analysis Uncovers Cell-Specific Regulatory Relationships in Embryonic, Juvenile, Adult, and Aged Mouse Lens Epithelium and Fibers
- PMID: 40238114
- PMCID: PMC12011134
- DOI: 10.1167/iovs.66.4.42
Transcriptome Meta-Analysis Uncovers Cell-Specific Regulatory Relationships in Embryonic, Juvenile, Adult, and Aged Mouse Lens Epithelium and Fibers
Abstract
Purpose: The lens transcriptome has been examined using microarrays and RNA-sequencing (RNA-seq). These omics data are the basis of the bioinformatics web-resource iSyTE that has identified new genes involved in lens development and cataract. The lens predominantly contains epithelial and fiber cells, and yet, presently, iSyTE is based on whole lens data. To gain cell-specific regulatory insights, we meta-analyzed isolated epithelium and fiber transcriptomes from embryonic/postnatal, adult and aged lenses.
Methods: Mouse lens epithelium and fiber transcriptome public datasets at embryonic (E) and postnatal (P) stages E12.5, E14.5, E16.5, E18.5, P0.5, P0, P5, P13, and age one month, three months, six months, and two years were analyzed. Microarray or RNA-seq data were analyzed by appropriate methods and compared to other resources (e.g., Cat-Map, CompBio).
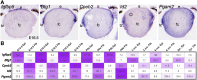
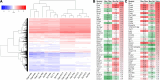
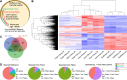
Results: Across all RNA-seq datasets examined, 2466 genes are differentially expressed between epithelium and fibers, of which 106 are cataract-linked. Gene ontology enrichment validates epithelial and fiber expression, corroborating the meta-analysis. Whole embryonic-body-in silico subtraction and other analyses identify several new high-priority epithelial- and/or fiber-enriched genes (e.g., Casz1, Ell2). Furthermore, new insights into cell-specific regulatory processes at distinct stages are identified (e.g., ribonucleoprotein regulation in E12.5 epithelium). Finally, this data is made accessible at iSyTE (https://research.bioinformatics.udel.edu/iSyTE/).
Conclusions: This spatiotemporal transcriptome meta-analysis comprehensively informs on epithelium- and fiber-specific regulatory processes in developing, adult and aged lenses. Notably, it includes the first description of an embryonic stage (i.e., E12.5) representing early primary fiber differentiation, thus informing on the initial transcriptome changes as lens cell-types are readily distinguishable.
Conflict of interest statement
Disclosure:
Figures








Similar articles
-
RNA sequencing-based transcriptomic profiles of embryonic lens development for cataract gene discovery.Hum Genet. 2018 Dec;137(11-12):941-954. doi: 10.1007/s00439-018-1958-0. Epub 2018 Nov 11. Hum Genet. 2018. PMID: 30417254 Free PMC article.
-
Molecular characterization of mouse lens epithelial cell lines and their suitability to study RNA granules and cataract associated genes.Exp Eye Res. 2015 Feb;131:42-55. doi: 10.1016/j.exer.2014.12.011. Epub 2014 Dec 19. Exp Eye Res. 2015. PMID: 25530357 Free PMC article.
-
Proteome-transcriptome analysis and proteome remodeling in mouse lens epithelium and fibers.Exp Eye Res. 2019 Feb;179:32-46. doi: 10.1016/j.exer.2018.10.011. Epub 2018 Oct 22. Exp Eye Res. 2019. PMID: 30359574 Free PMC article.
-
Systems biology of lens development: A paradigm for disease gene discovery in the eye.Exp Eye Res. 2017 Mar;156:22-33. doi: 10.1016/j.exer.2016.03.010. Epub 2016 Mar 16. Exp Eye Res. 2017. PMID: 26992779 Free PMC article. Review.
-
RNA-binding proteins and post-transcriptional regulation in lens biology and cataract: Mediating spatiotemporal expression of key factors that control the cell cycle, transcription, cytoskeleton and transparency.Exp Eye Res. 2022 Jan;214:108889. doi: 10.1016/j.exer.2021.108889. Epub 2021 Dec 11. Exp Eye Res. 2022. PMID: 34906599 Free PMC article. Review.
References
-
- Chauhan BK, Reed NA, Zhang W, Duncan MK, Kilimann MW, Cvekl A.. Identification of genes downstream of Pax6 in the mouse lens using cDNA microarrays. J Biol Chem. 2002; 277: 11539–11548. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

