Amyotrophic lateral sclerosis and frontotemporal dementia mutation reduces endothelial TDP-43 and causes blood-brain barrier defects
- PMID: 40238886
- PMCID: PMC12002129
- DOI: 10.1126/sciadv.ads0505
Amyotrophic lateral sclerosis and frontotemporal dementia mutation reduces endothelial TDP-43 and causes blood-brain barrier defects
Abstract
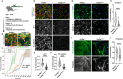
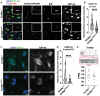
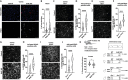
Mutations in the TARDBP gene encoding TDP-43 protein are linked to loss of function in neurons and familial frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS). We recently identified reduced nuclear TDP-43 in capillary endothelial cells (ECs) of donors with ALS-FTD. Because blood-brain barrier (BBB) permeability increases in ALS-FTD, we postulated that reduced nuclear TDP-43 in ECs might contribute. Here, we show that nuclear TDP-43 is reduced in ECs of mice with an ALS-FTD-associated mutation in TDP-43 (TardbpG348C) and that this leads to cell-autonomous loss of junctional complexes and BBB integrity. Targeted excision of TDP-43 in brain ECs recapitulates BBB defects and loss of junctional complexes and ultimately leads to fibrin deposition, gliosis, phospho-Tau accumulation, and impaired memory and social interaction. Transcriptional changes in TDP-43-deficient ECs resemble diseased brain ECs. These data show that nuclear loss of TDP-43 in brain ECs disrupts the BBB and causes hallmarks of FTD.
Figures




Update of
-
Loss of Endothelial TDP-43 Leads to Blood Brain Barrier Defects in Mouse Models of Amyotrophic Lateral Sclerosis and Frontotemporal Dementia.bioRxiv [Preprint]. 2023 Dec 14:2023.12.13.571184. doi: 10.1101/2023.12.13.571184. bioRxiv. 2023. Update in: Sci Adv. 2025 Apr 18;11(16):eads0505. doi: 10.1126/sciadv.ads0505. PMID: 38168388 Free PMC article. Updated. Preprint.
References
-
- Tziortzouda P., Van Den Bosch L., Hirth F., Triad of TDP43 control in neurodegeneration: Autoregulation, localization and aggregation. Nat. Rev. Neurosci. 22, 197–208 (2021). - PubMed
-
- Brown A.-L., Wilkins O. G., Keuss M. J., Hill S. E., Zanovello M., Lee W. C., Bampton A., Lee F. C. Y., Masino L., Qi Y. A., Bryce-Smith S., Gatt A., Hallegger M., Fagegaltier D., Phatnani H., NYGC ALS Consortium, Newcombe J., Gustavsson E. K., Seddighi S., Reyes J. F., Coon S. L., Ramos D., Schiavo G., Fisher E. M. C., Raj T., Secrier M., Lashley T., Ule J., Buratti E., Humphrey J., Ward M. E., Fratta P., TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature 603, 131–137 (2022). - PMC - PubMed
-
- Ma X. R., Prudencio M., Koike Y., Vatsavayai S. C., Kim G., Harbinski F., Briner A., Rodriguez C. M., Guo C., Akiyama T., Schmidt H. B., Cummings B. B., Wyatt D. W., Kurylo K., Miller G., Mekhoubad S., Sallee N., Mekonnen G., Ganser L., Rubien J. D., Jansen-West K., Cook C. N., Pickles S., Oskarsson B., Graff-Radford N. R., Boeve B. F., Knopman D. S., Petersen R. C., Dickson D. W., Shorter J., Myong S., Green E. M., Seeley W. W., Petrucelli L., Gitler A. D., TDP-43 represses cryptic exon inclusion in the FTD-ALS gene UNC13A. Nature 603, 124–130 (2022). - PMC - PubMed
-
- Melamed Z., López-Erauskin J., Baughn M. W., Zhang O., Drenner K., Sun Y., Freyermuth F., McMahon M. A., Beccari M. S., Artates J. W., Ohkubo T., Rodriguez M., Lin N., Wu D., Bennett C. F., Rigo F., Da Cruz S., Ravits J., Lagier-Tourenne C., Cleveland D. W., Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat. Neurosci. 22, 180–190 (2019). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

