Mitochondrial metabolism sustains DNMT3A-R882-mutant clonal haematopoiesis
- PMID: 40239706
- PMCID: PMC12158785
- DOI: 10.1038/s41586-025-08980-6
Mitochondrial metabolism sustains DNMT3A-R882-mutant clonal haematopoiesis
Abstract
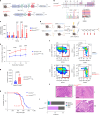
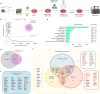
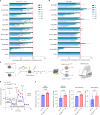
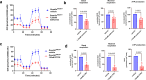
Somatic DNMT3A-R882 codon mutations drive the most common form of clonal haematopoiesis (CH) and are associated with increased acute myeloid leukaemia (AML) risk1,2. Preventing expansion of DNMT3A-R882-mutant haematopoietic stem/progenitor cells (HSPCs) may therefore avert progression to AML. To identify DNMT3A-R882-mutant-specific vulnerabilities, we conducted a genome-wide CRISPR screen on primary mouse Dnmt3aR882H/+ HSPCs. Among the 640 vulnerability genes identified, many were involved in mitochondrial metabolism, and metabolic flux analysis confirmed enhanced oxidative phosphorylation use in Dnmt3aR882H/+ versus Dnmt3a+/+ (WT) HSPCs. We selected citrate/malate transporter Slc25a1 and complex I component Ndufb11, for which pharmacological inhibitors are available, for downstream studies. In vivo administration of SLC25A1 inhibitor CTPI2 and complex I inhibitors IACS-010759 and metformin suppressed post-transplantation clonal expansion of Dnmt3aR882H/+, but not WT, long-term haematopoietic stem cells. The effect of metformin was recapitulated using a primary human DNMT3A-R882 CH sample. Notably, analysis of 412,234 UK Biobank participants showed that individuals taking metformin had a markedly lower prevalence of DNMT3A-R882-mutant CH, after controlling for potential confounders including glycated haemoglobin, diabetes and body mass index. Collectively, our data propose modulation of mitochondrial metabolism as a therapeutic strategy for prevention of DNMT3A-R882-mutant AML.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: G.S.V. is a consultant for STRM.BIO and receives a research grant from AstraZeneca. S.W., J.M. and M.A.F. are current employees and/or stockholders of AstraZeneca. K.T. has received consultancy fees, stock options and research funding from Storm Therapeutics Ltd., Cambridge, UK. K.T. is a cofounder of Btwo3 Therapeutics LLC, USA and TEP Therapeutics LLC, USA. The other authors declare no competing interests.
Figures










References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

