Molecular basis for RNA discrimination by human DNA ligase 1
- PMID: 40239996
- PMCID: PMC12000876
- DOI: 10.1093/nar/gkaf299
Molecular basis for RNA discrimination by human DNA ligase 1
Abstract
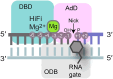
DNA ligase 1 (LIG1) finalizes DNA replication and repair by catalyzing the joining of DNA nicks. LIG1 is highly specific for DNA-DNA junctions over DNA-RNA junctions, discriminating strongly against a single ribonucleotide at the 5' side of the nick. This selectivity of LIG1 prevents futile and potentially mutagenic DNA-RNA cleavage and re-ligation cycles during Okazaki fragment maturation or ribonucleotide excision repair of genome-embedded ribonucleotide monophosphates (rNMPs), but the determinants of LIG1 rNMP discrimination are ill-defined. We report structural and kinetic analysis of LIG1 DNA-RNA complexes showing that LIG1 employs an aromatic steric gate to stabilize the enzyme-substrate complex and directly exclude rNMP-containing polynucleotides. Mutation of this RNA gate compromises the adenylyl-transfer and nick-sealing reactions but decreases the discrimination against an rNMP-containing substrate by ∼3600-fold. Our results establish the role of the conserved steric gate in ribonucleotide discrimination by high-fidelity (HiFi) DNA ligases at each step of the ligation reaction, which has parallels to the ribonucleotide discrimination by HiFi DNA polymerases.
Published by Oxford University Press on behalf of Nucleic Acids Research 2025.
Conflict of interest statement
None declared.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

