Genome-wide association meta-analyses of drug-resistant epilepsy
- PMID: 40240269
- PMCID: PMC12143209
- DOI: 10.1016/j.ebiom.2025.105675
Genome-wide association meta-analyses of drug-resistant epilepsy
Abstract
Background: Epilepsy is one of the most common neurological disorders, affecting over 50 million people worldwide. One-third of people with epilepsy do not respond to currently available anti-seizure medications, constituting one of the most important problems in epilepsy. Little is known about the molecular pathology of drug resistance in epilepsy, in particular, possible underlying genetic factors are largely unknown.
Methods: We performed a genome-wide association study (GWAS) in two epilepsy cohorts of European ancestry, comparing drug-resistant (N = 4208) to drug-responsive individuals (N = 2618) followed by meta-analyses across the studies. Next, we performed subanalyses split into two broad subtypes: acquired or non-acquired focal and genetic generalized epilepsy.
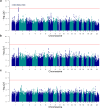
Findings: Our drug-resistant versus drug-responsive epilepsy GWAS meta-analysis showed no significant loci when combining all epilepsy types. Sub-analyses on individuals with focal epilepsy (FE) identified a significant locus on chromosome 1q42.11-q42.12 (lead SNP: rs35915186, P = 1·51 × 10-8, OR[C] = 0·74). This locus was not associated with any epilepsy subtype in the latest epilepsy GWAS (lowest uncorrected P = 0·009 for FE vs. healthy controls), and drug resistance in FE was not genetically correlated with susceptibility to FE itself. Seven genome-wide significant SNPs within this locus, encompassing the genes CNIH4, WDR26, and CNIH3, were identified to protect against drug-resistant FE. Further transcriptome-wide association studies (TWAS) imply significantly higher expression levels of CNIH3 and WDR26 in drug-resistant FE than in drug-responsive FE. CNIH3 is implicated in AMPA receptor assembly and function, while WDR26 haploinsufficiency is linked to intellectual disability and seizures. These findings suggest that CNIH3 and WDR26 may play a role in mediating drug response in focal epilepsy.
Interpretation: We identified a contribution of common genetic variation to drug-resistant focal epilepsy. These findings provide insights into possible mechanisms underlying drug response variability in epilepsy, offering potential targets for personalised treatment approaches.
Funding: This work is part of the European Union's Seventh Framework Programme (FP7/2007-2013) under grant agreement n° 279062 (EpiPGX) and the Centers for Common Disease Genomics (CCDG) program, funded by the National Human Genome Research Institute (NHGRI) and the National Heart, Lung, and Blood Institute (NHLBI).
Keywords: Antiseizure medication; Association; Genetics; Pharmacogenomics; Treatment.
Copyright © 2025. Published by Elsevier B.V.
Conflict of interest statement
Declaration of interests AA is an employee of Regenon and owns Regenon stocks. AGM received institutional consulting fees from Jazz Pharmaceuticals and UCB Pharma; institutional honoraria for lectures from Sanofi and GSK, institutional support for meeting attendance/travel from Angelini Pharma, and has unpaid leadership roles at the European Academy of Neurology and Epilepsy Research Institute. BMN is a member of the Neumora Scientific and Deep Genomics Scientific Advisory boards. CD received institutional honoraria from UCB Pharma, support for meeting attendance/travel from Angelini Pharma, and is a member of Angelini Pharma and Neuraxpharm Advisory Boards. DL received institutional research funding from the National Institutes of Health (NIH), National Institute of Neurological Disorders and Stroke (NINDS) under R01 NS117544. GC received research funding from Janssen Pharmaceuticals and consultancy fees from Ono Pharmaceuticals. GJS received personal consulting fees from Angelini Pharma, and personal honoraria for lectures from Angelini Pharma, Bial Pharma UK, and UCB Pharma. HL received personal consulting fees from Praxis Precision Medicine and institutional fees from Lario Therapeutics, personal lecture honoraria from Eisai and UCB Pharma, personal payment for expert testimony for Fondazione Telethon, and is an advisory board member of IntraBio. HS, KS, and UU are employees of deCODE genetics/Amgen Inc. JJC received honoraria from UCB-Pharma, Glaxo Smith Kline, Janssen-Cilag, Sanofi-Synthelabo, Pfizer, and Eisai to attend advisory boards, present lectures/tutorials. JWS received institutional research funding from the UK National Institute for Health Research, Angelini Pharma, UCB Pharma, Epilepsy NL, the Academy of Medical Sciences, and the Wellcome Trust Alliance, institutional consulting fees from the UK Competition & Markets Authority and Eisai, personal honoraria from Angelini Pharma, Eisai, and UCB, and personal fees for participation on an advisory board of Angelini Pharma, and holds unpaid roles as Medical Director of the UK Epilepsy Society and Editorial Board member for Lancet Neurology. LJ received institutional research funding from the Swedish state under the ALF agreement (ALFGBG-966370), Genomic Medicine Sweden (GMS K131050263), and the Söderström König Foundation. MRJ received research funding from the UK Research and Innovation (UKRI) Medical Research Council (MRC) (Award Nos. MR/S02638X/1 and MR/W029790/1). ND received consulting fees and honoraria from Angelini Pharma, Actiobio, Eisai, UNEEG Medical, and Jazz Pharma, and owns stocks from Actiobio. PS received consulting fees from Jazz Healthcare and UCB Pharma for participation on advisory boards, and honoraria from Proveca and UCB Pharma for presentation at congresses/workshops. SFB received institutional research funding from the National Health and Medical Research Council (NHMRC) (Grant IDs 1091593, 1196637, 2010562), UCB Pharma, Eisai, SEER, Chiesi, and LivaNova, personal consulting fees from Praxis Precision Medicines, personal honoraria from Eisai and DeltaMed, co-owns a patent held by Bionomics Inc. licensed to Athena Diagnostics, Genetics Technologies Ltd, and is the Chief Medical Officer of the Epilepsy Foundation (Victoria). SMS received institutional research funding from CURE Epilepsy, Epilepsy Society, and MRC, honoraria from Angelini Pharma, Eisai, Zogenix, UCB, Eisai, Jazz Pharmaceuticals, and UCB, travel support from UCB Pharma, and is a member of Advisory Boards of Biocodex, Stoke Therapeutics, and Takeda. TJOB received institutional research funding from NHMRC, MRFF, DOD, and NIH, institutional consulting fees from Kinoxis Therapeutics, Jazz Pharmaceuticals, and Livanova, institutional honoraria from UCB, institutional travel support from Longboard Pharmaceuticals, is a member of the Kinoxis Therapeutics Advisory Board, and hold an unpaid position as Cabrini Health Board Member. All other authors have no conflict of interests to declare.
Figures

References
-
- Bell G.S., Neligan A., Sander J.W. An unknown quantity--the worldwide prevalence of epilepsy. Epilepsia. 2014;55(7):958–962. - PubMed
-
- Kwan P., Arzimanoglou A., Berg A.T., et al. Definition of drug resistant epilepsy: consensus proposal by the ad hoc Task Force of the ILAE Commission on Therapeutic Strategies. Epilepsia. 2010;51(6):1069–1077. - PubMed
-
- Tellez-Zenteno J.F., Patten S.B., Jetté N., Williams J., Wiebe S. Psychiatric comorbidity in epilepsy: a population-based analysis. Epilepsia. 2007;48(12):2336–2344. - PubMed
-
- Téllez-Zenteno J.F., Matijevic S., Wiebe S. Somatic comorbidity of epilepsy in the general population in Canada. Epilepsia. 2005;46(12):1955–1962. - PubMed
-
- Lang J., Jeschke S., Herziger B., et al. Prejudices against people with epilepsy as perceived by affected people and their families. Epilepsy Behav. 2022;127 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

