Inflammatory conditions shape phenotypic and functional characteristics of lung-resident memory T cells in mice
- PMID: 40240341
- PMCID: PMC12003732
- DOI: 10.1038/s41467-025-58931-y
Inflammatory conditions shape phenotypic and functional characteristics of lung-resident memory T cells in mice
Abstract
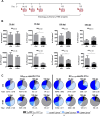
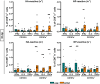
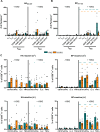
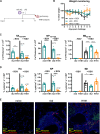
Lung tissue-resident memory T cells (TRM) are critical for the local control of respiratory tract infections caused by influenza A viruses (IAV). Here we compare TRM populations induced by intranasal adenoviral vector vaccines encoding hemagglutinin and nucleoprotein (NP) with those induced by an H1N1 infection in BALB/c mice. While vaccine-induced TRM express high levels of CD103 and persist longer in the lung parenchyma, short-lived, H1N1-induced TRM have a transcriptome associated with higher cytotoxic potential and distinct transcriptional profile as shown by single-cell RNA sequencing. In both the vaccine and H1N1 groups, NP-specific CD8+ T cells expand during heterologous influenza virus infection and protect the mice from disease. Meanwhile, lung inflammation in response to an infection with unrelated respiratory syncytial virus do not influence the fate of pre-existing TRM. Our preclinical work thus confirms that inflammatory conditions in the tissue shape the phenotypic and functional characteristics of TRM to serve relevant informations for optimizing mucosal vaccines.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- WHO. Influenza (Seasonal). https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (2025).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

