Multi-zonal liver organoids from human pluripotent stem cells
- PMID: 40240593
- PMCID: PMC12488051
- DOI: 10.1038/s41586-025-08850-1
Multi-zonal liver organoids from human pluripotent stem cells
Erratum in
-
Publisher Correction: Multi-zonal liver organoids from human pluripotent stem cells.Nature. 2025 Jun;642(8067):E16. doi: 10.1038/s41586-025-09123-7. Nature. 2025. PMID: 40404938 No abstract available.
Abstract
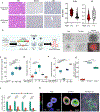
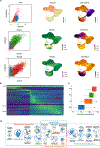
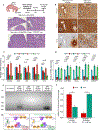
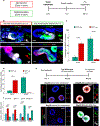
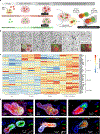
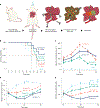
Distinct hepatocyte subpopulations are spatially segregated along the portal-central axis and are critical to understanding metabolic homeostasis and injury in the liver1. Although several bioactive molecules, including ascorbate and bilirubin, have been described as having a role in directing zonal fates, zonal liver architecture has not yet been replicated in vitro2,3. Here, to evaluate hepatic zonal polarity, we developed a self-assembling zone-specific liver organoid by co-culturing ascorbate- and bilirubin-enriched hepatic progenitors derived from human induced pluripotent stem cells. We found that preconditioned hepatocyte-like cells exhibited zone-specific functions associated with the urea cycle, glutathione synthesis and glutamate synthesis. Single-nucleus RNA-sequencing analysis of these zonally patterned organoids identifies a hepatoblast differentiation trajectory that dictates periportal, interzonal and pericentral human hepatocytes. Epigenetic and transcriptomic analysis showed that zonal identity is orchestrated by ascorbate- or bilirubin-dependent binding of EP300 to TET1 or HIF1α. Transplantation of the self-assembled zonally patterned human organoids improved survival of immunodeficient rats who underwent bile duct ligation by ameliorating the hyperammonaemia and hyperbilirubinaemia. Overall, this multi-zonal organoid system serves as an in vitro human model to better recapitulate hepatic architecture relevant to liver development and disease.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures











Update of
-
Self-Assembled Generation of Multi-zonal Liver Organoids from Human Pluripotent Stem Cells.bioRxiv [Preprint]. 2024 Aug 30:2024.08.30.610426. doi: 10.1101/2024.08.30.610426. bioRxiv. 2024. Update in: Nature. 2025 May;641(8065):1258-1267. doi: 10.1038/s41586-025-08850-1. PMID: 39257824 Free PMC article. Updated. Preprint.
References
-
- Ben-Moshe S & Itzkovitz S Spatial heterogeneity in the mammalian liver. Nat. Rev. Gastroenterol. Hepatol. 16, 395–410 (2019). - PubMed
-
- Martini T, Naef F & Tchorz JS Spatiotemporal metabolic liver zonation and consequences on pathophysiology. Annu. Rev. Pathol. Mech. Dis. 18, 439–466 (2023). - PubMed
-
- Bartl M et al. Optimality in the zonation of ammonia detoxification in rodent liver. Arch. Toxicol. 89, 2069–2078 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

