Histone H1 deamidation facilitates chromatin relaxation for DNA repair
- PMID: 40240600
- PMCID: PMC12074999
- DOI: 10.1038/s41586-025-08835-0
Histone H1 deamidation facilitates chromatin relaxation for DNA repair
Abstract
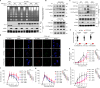
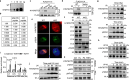
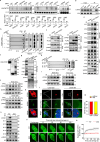
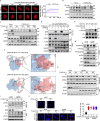
The formation of accessible chromatin around DNA double-strand breaks is essential for their efficient repair1. Although the linker histone H1 is known to facilitate higher-order chromatin compaction2,3, the mechanisms by which H1 modifications regulate chromatin relaxation in response to DNA damage are unclear. Here we show that CTP synthase 1 (CTPS1)-catalysed deamidation of H1 asparagine residues 76 and 77 triggers the sequential acetylation of lysine 75 following DNA damage, and this dual modification of H1 is associated with chromatin opening. Mechanistically, the histone acetyltransferase p300 showed a preference for deamidated H1 as a substrate, establishing H1 deamidation as a prerequisite for subsequent acetylation. Moreover, high expression of CTPS1 was associated with resistance to cancer radiotherapy, in both mouse xenograft models and clinical cohorts. These findings provide new insights into how linker histones regulate dynamic chromatin alterations in the DNA damage response.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures











References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

