Structure of the ATP-driven methyl-coenzyme M reductase activation complex
- PMID: 40240609
- PMCID: PMC12176620
- DOI: 10.1038/s41586-025-08890-7
Structure of the ATP-driven methyl-coenzyme M reductase activation complex
Abstract
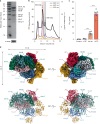
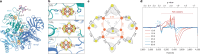
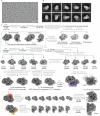
Methyl-coenzyme M reductase (MCR) is the enzyme responsible for nearly all biologically generated methane1. Its active site comprises coenzyme F430, a porphyrin-based cofactor with a central nickel ion that is active exclusively in the Ni(I) state2,3. How methanogenic archaea perform the reductive activation of F430 represents a major gap in our understanding of one of the most ancient bioenergetic systems in nature. Here we purified and characterized the MCR activation complex from Methanococcus maripaludis. McrC, a small subunit encoded in the mcr operon, co-purifies with the methanogenic marker proteins Mmp7, Mmp17, Mmp3 and the A2 component. We demonstrated that this complex can activate MCR in vitro in a strictly ATP-dependent manner, enabling the formation of methane. In addition, we determined the cryo-electron microscopy structure of the MCR activation complex exhibiting different functional states with local resolutions reaching 1.8-2.1 Å. Our data revealed three complex iron-sulfur clusters that formed an electron transfer pathway towards F430. Topology and electron paramagnetic resonance spectroscopy analyses indicate that these clusters are similar to the [8Fe-9S-C] cluster, a maturation intermediate of the catalytic cofactor in nitrogenase. Altogether, our findings offer insights into the activation mechanism of MCR and prospects on the early evolution of nitrogenase.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures










References
-
- Jackson, R. B. et al. Human activities now fuel two-thirds of global methane emissions. Environ. Res. Lett.19, 101002 (2024).
-
- Ermler, U., Grabarse, W., Shima, S., Goubeaud, M. & Thauer, R. K. Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation. Science278, 1457–1462 (1997). - PubMed
-
- Grabarse, W. et al. On the mechanism of biological methane formation: structural evidence for conformational changes in methyl-coenzyme M reductase upon substrate binding. J. Mol. Biol.309, 315–330 (2001). - PubMed
-
- Evans, P. N. et al. An evolving view of methane metabolism in the Archaea. Nat. Rev. Microbiol.17, 219–232 (2019). - PubMed
-
- Scheller, S., Goenrich, M., Boecher, R., Thauer, R. K. & Jaun, B. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature465, 606–608 (2010). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

