The de novo DNA methyltransferase 3B is a novel epigenetic regulator of MYC in multiple myeloma, representing a promising therapeutic target to counter relapse
- PMID: 40241199
- PMCID: PMC12004749
- DOI: 10.1186/s13046-025-03382-y
The de novo DNA methyltransferase 3B is a novel epigenetic regulator of MYC in multiple myeloma, representing a promising therapeutic target to counter relapse
Abstract
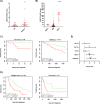
Background: The plasma cell malignancy multiple myeloma (MM) remains incurable due to the inevitable development of drug resistance (DR). Epigenetic modifiers are frequently mutated or deregulated in MM patients, contributing to MM progression and relapse. Overexpression of the de novo DNA methyltransferase 3B (DNMT3B) in MM has been reported, correlating with poor prognosis. However, its exact role in MM cell biology and relapse remains elusive.
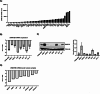
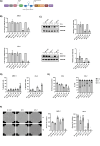
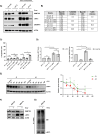
Methods: To evaluate the basal expression and prognostic value of DNMT3B mRNA in terms of overall survival the publicly available gene expression profiling datasets GSE2658, GSE9782, GSE4581, E-MTAB-372, E-TABM-1088 and E-TABM-937 were used. Both the DNMT3B selective inhibitor Nanaomycin A and genetic knockdown using a doxycycline inducible shRNA against DNMT3B were used to target DNMT3B. Viability and apoptosis were assessed using respectively a CellTiter-Glo assay and AnnexinV/7AAD stainings. Cell proliferation was measured by BrdU incorporation and cell cycle analysis, while the clonogenic capacity was evaluated by a colony formation assay. Finally, RNA-seq was performed upon genetic knockdown.
Results: Here, we show that DNMT3B is significantly increased in the relapsed setting and high DNMT3B levels are strongly correlating with disease progression and high-risk disease, irrespective of the treatment. Targeting DNMT3B using either genetic inhibition or the selective inhibitor Nanaomycin A strongly impaired MM cell growth, survival and clonogenicity. Moreover, Nanaomycin A reduced viability of primary MM cells from newly diagnosed and relapsed patients. Mechanistic studies revealed that DNMT3B inhibition mainly affects cell cycle and stemness-related transcriptional programs. Notably, DNMT3B depletion affected the stability of the master cell cycle regulator MYC, thereby reducing c-MYC levels and cell viability both in parental and c-MYC overexpressing cells. Finally, Nanaomycin A (re)sensitized MM cells to bortezomib, melphalan and anti-CD38 monoclonal antibodies (daratumumab, isatuximab).
Conclusion: Collectively, our findings uncover DNMT3B as a targetable vulnerability in high-risk patients with high DNMT3B/MYC levels.
Keywords: DNMT3B; Epigenetics; Multiple myeloma; Relapse.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: BM samples from patients were collected for routine diagnostic or evaluation purposes after patients’ written informed consent was given and in accordance with the Declaration of Helsinki and institutional research board approval from Brussels University hospital (B.U.N. 143201838414) and Montpellier University hospital (DC2008-417). Peripheral mononuclear cells (PBMC) of healthy donors for NK isolation were obtained from the Hematology unit of the university hospital (UZ Brussels), after informed consent, conform with the declaration of Helsinki. In vivo experiments in mice were approved by the VUB Ethical committee (Licence No LA1230281, CEP No 23-281-13). Consent for publication: All authors read and approved the final manuscript. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
The DNMT3B Inhibitor Nanaomycin A as a Neuroblastoma Therapeutic Agent.Curr Cancer Drug Targets. 2023;23(11):837-842. doi: 10.2174/1568009623666230522113645. Curr Cancer Drug Targets. 2023. PMID: 37221685
-
DNMT3b/OCT4 expression confers sorafenib resistance and poor prognosis of hepatocellular carcinoma through IL-6/STAT3 regulation.J Exp Clin Cancer Res. 2019 Nov 26;38(1):474. doi: 10.1186/s13046-019-1442-2. J Exp Clin Cancer Res. 2019. PMID: 31771617 Free PMC article.
-
Targeting Acetyl-CoA Carboxylase Suppresses De Novo Lipogenesis and Tumor Cell Growth in Multiple Myeloma.Clin Cancer Res. 2025 May 15;31(10):1975-1987. doi: 10.1158/1078-0432.CCR-24-2000. Clin Cancer Res. 2025. PMID: 40053701
-
Methyltransferase DNMT3B in leukemia.Leuk Lymphoma. 2020 Feb;61(2):263-273. doi: 10.1080/10428194.2019.1666377. Epub 2019 Sep 24. Leuk Lymphoma. 2020. PMID: 31547729 Review.
-
Using human disease mutations to understand de novo DNA methyltransferase function.Biochem Soc Trans. 2024 Oct 30;52(5):2059-2075. doi: 10.1042/BST20231017. Biochem Soc Trans. 2024. PMID: 39446312 Free PMC article. Review.
Cited by
-
Unveiling the role of NAD glycohydrolase CD38 in aging and age-related diseases: insights from bibliometric analysis and comprehensive review.Front Immunol. 2025 Jun 2;16:1579924. doi: 10.3389/fimmu.2025.1579924. eCollection 2025. Front Immunol. 2025. PMID: 40529366 Free PMC article.
-
CXCR Family and Hematologic Malignancies in the Bone Marrow Microenvironment.Biomolecules. 2025 May 13;15(5):716. doi: 10.3390/biom15050716. Biomolecules. 2025. PMID: 40427609 Free PMC article. Review.
References
-
- Galm O, et al. DNA methylation changes in multiple myeloma. Leukemia. 2004;18:1687–92. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

