Coupling deep phenotypic quantification with next-generation phenotyping for 192 individuals with germline histonopathies
- PMID: 40241305
- PMCID: PMC12142524
- DOI: 10.1016/j.xhgg.2025.100440
Coupling deep phenotypic quantification with next-generation phenotyping for 192 individuals with germline histonopathies
Abstract
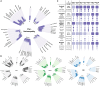
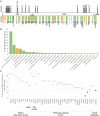
Mendelian histonopathies are rare neurodevelopmental disorders (NDDs) caused by germline variants in histone-encoding genes. Here, we perform a more expansive pan-histonopathy interrogation than previously possible. We analyze data from 192 individuals affected by histonopathies. This analysis includes representation of the 185 published individuals with HIST1H1E syndrome, Bryant-Li-Bhoj syndrome, and Tessadori-Bicknell-van Haaften NDD; as well as from seven unpublished individuals, five of whom harbor variants in genes not previously associated with disease (HIST1H2AL/H2AC16, H2AFZ/H2AZ1, HIST1H3D/H3C4, and HIST3H3/H3-4). By intersecting clinician-reported phenotypic data with next-generation phenotyping of published 2D facial photographs (n = 98), we sought to address the lack of established craniofacial gestalts or characteristic phenotypic patterns for this community. While these analyses may suggest a histone core versus linker protein basis of delineation, they more strikingly highlight data gaps that confound the identification of phenotypic patterns at this time. Based on this, we developed an updated standardized clinical survey, which allowed us to identify the second known individual with a germline histonopathy and a cancer diagnosis. Notably, the community-wide cancer incidence is currently 1%, which falls below the recommended 5% cut off for routine surveillance. Ultimately, this work highlights the ways in which histonopathy-associated phenotypes change throughout the lifespan, necessitating longitudinal re-evaluation; that every identified individual shapes our understanding of these syndromes in a way that improves care for this community; and the value of ongoing translational work to address the outstanding question of cancer predisposition for individuals living with germline histonopathies.
Keywords: Mendelian genetics; comorbidity; histones; neurodevelopmental disorders; next-generation phenotyping; translational genetics.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
MeSH terms
LinkOut - more resources
Full Text Sources

