CRISPR/Cas9-mediated SHP-1-knockout T cells combined with simvastatin enhances anti-tumor activity in humanized-PDX HCC model
- PMID: 40241752
- PMCID: PMC12003012
- DOI: 10.1016/j.isci.2025.112266
CRISPR/Cas9-mediated SHP-1-knockout T cells combined with simvastatin enhances anti-tumor activity in humanized-PDX HCC model
Abstract
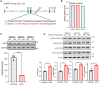
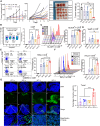
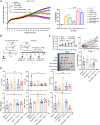
Hepatocellular carcinoma (HCC) resists immunotherapy due to its immunosuppressive microenvironment. Sarcoma homology 2 domain-containing protein tyrosine phosphatase-1 (SHP-1) inhibits T cell receptor signaling, and its pharmacological inhibition is limited by poor selectivity and membrane permeability. Here, we generated CRISPR-edited SHP-1-knockout (KO) CD8+ T cells to enhance adoptive therapy against HCC. Single-cell RNA sequencing of HCC patient T cells revealed elevated SHP-1 in exhausted subsets. SHP-1-KO T cells exhibited increased effector memory T cells (TEM) proportions and enhanced IFN-γ/Granzyme B/perforin secretion, improving cytotoxicity against HCC lines. In humanized PDX models, SHP-1-KO T cells demonstrated superior tumor-killing activity. Transcriptomics identified upregulated lipid metabolism pathways, with HMGCR as a hub gene. Combining SHP-1-KO T cells with simvastatin (HMGCR inhibitor) synergistically amplified anti-HCC efficacy. This study proposes a dual strategy combining SHP-1-targeted cell therapy and metabolic modulation to overcome immunotherapy resistance, offering a translatable approach for HCC treatment.
Keywords: Biological sciences; Cancer; Cancer systems biology; Natural sciences; Systems biology.
© 2025 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Abrogation of SRC homology region 2 domain-containing phosphatase 1 in tumor-specific T cells improves efficacy of adoptive immunotherapy by enhancing the effector function and accumulation of short-lived effector T cells in vivo.J Immunol. 2012 Aug 15;189(4):1812-25. doi: 10.4049/jimmunol.1200552. Epub 2012 Jul 13. J Immunol. 2012. PMID: 22798667 Free PMC article.
-
CRISPR/Cas9-mediated knockout of intracellular molecule SHP-1 enhances tumor-killing ability of CD133-targeted CAR T cells in vitro.Exp Hematol Oncol. 2023 Oct 6;12(1):88. doi: 10.1186/s40164-023-00450-x. Exp Hematol Oncol. 2023. PMID: 37803455 Free PMC article.
-
The HNF1α-regulated lncRNA HNF1A-AS1 reverses the malignancy of hepatocellular carcinoma by enhancing the phosphatase activity of SHP-1.Mol Cancer. 2018 Feb 21;17(1):63. doi: 10.1186/s12943-018-0813-1. Mol Cancer. 2018. PMID: 29466992 Free PMC article.
-
New and Unexpected Biological Functions for the Src-Homology 2 Domain-Containing Phosphatase SHP-2 in the Gastrointestinal Tract.Cell Mol Gastroenterol Hepatol. 2015 Nov 14;2(1):11-21. doi: 10.1016/j.jcmgh.2015.11.001. eCollection 2016 Jan. Cell Mol Gastroenterol Hepatol. 2015. PMID: 28174704 Free PMC article. Review.
-
Optimizing cancer treatment: the synergistic potential of CAR-T cell therapy and CRISPR/Cas9.Front Immunol. 2024 Nov 8;15:1462697. doi: 10.3389/fimmu.2024.1462697. eCollection 2024. Front Immunol. 2024. PMID: 39582866 Free PMC article. Review.
Cited by
-
Multi-omics dissection of CPNE1 reveals its prognostic value and immune-regulatory function in liver cancer.Discov Oncol. 2025 Aug 7;16(1):1499. doi: 10.1007/s12672-025-03366-6. Discov Oncol. 2025. PMID: 40775545 Free PMC article.
References
-
- Wolchok J.D., Neyns B., Linette G., Negrier S., Lutzky J., Thomas L., Waterfield W., Schadendorf D., Smylie M., Guthrie T., Jr., et al. Ipilimumab monotherapy in patients with pretreated advanced melanoma: a randomised, double-blind, multicentre, phase 2, dose-ranging study. Lancet Oncol. 2010;11:155–164. doi: 10.1016/S1470-2045(09)70334-1. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

