Pangenome analysis indicates evolutionary origins and genetic diversity: emphasis on the role of nodulation in symbiotic Bradyrhizobium
- PMID: 40241821
- PMCID: PMC12000093
- DOI: 10.3389/fpls.2025.1539151
Pangenome analysis indicates evolutionary origins and genetic diversity: emphasis on the role of nodulation in symbiotic Bradyrhizobium
Abstract
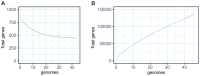
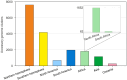
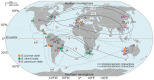
The Bradyrhizobium genus is widely known for encompassing many species capable of forming nodules and establishing the biological nitrogen fixation process with several legumes, significantly contributing to agriculture and environmental sustainability. Despite its importance, questions about the evolution, pangenome, and symbiotic genes of Bradyrhizobium are still poorly understood. In this study, we analyzed the pangenome of a set of Bradyrhizobium symbiotic species using the Roary and GET_HOMOLOGUES tools in strains originated from the Northern and Southern Hemispheres. We also investigated the presence and correlation of the fix, nif, nod, Type III secretion system (T3SS) and their effector proteins, and T4SS genes, trying to find differences between clades, hosts, and biogeographic origin. Pangenome analysis of Bradyrhizobium species from the Northern and Southern Hemispheres provided valuable insights into their diversity, biogeography, origin, and co-evolution with their legume host plants. The genus possesses a relatively small core genome compared to the expanded accessory genome, a key feature that facilitates genetic exchange and acquisition of new genes, allowing adaptation to a variety of environments. Notably, the presence or absence of T3SS effector proteins varied significantly according to the geographic location, suggesting specific environmental adaptations, as well as a direct relationship with nodulation genes. Comparative analysis indicated that symbiotic Bradyrhizobium species originated in the Northern Hemisphere and present a greater diversity of orthologous groups than those from the Southern Hemisphere. These results contribute to our understanding of the evolutionary history of these symbiotic bacteria.
Keywords: Bradyrhizobium; Fix/Nif; Nod factors; biogeography; environmental adaptation; nodulation; pangenome; type III secretion system.
Copyright © 2025 Terra, Klepa, Nogueira and Hungria.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Phylogeny of nodulation and nitrogen-fixation genes in Bradyrhizobium: supporting evidence for the theory of monophyletic origin, and spread and maintenance by both horizontal and vertical transfer.Int J Syst Evol Microbiol. 2011 Dec;61(Pt 12):3052-3067. doi: 10.1099/ijs.0.028803-0. Epub 2011 Feb 25. Int J Syst Evol Microbiol. 2011. PMID: 21357454
-
Phylogenetic distribution and evolutionary dynamics of nod and T3SS genes in the genus Bradyrhizobium.Microb Genom. 2020 Sep;6(9):mgen000407. doi: 10.1099/mgen.0.000407. Epub 2020 Aug 12. Microb Genom. 2020. PMID: 32783800 Free PMC article.
-
Symbiosis Contribution of Non-nodulating Bradyrhizobium cosmicum S23321 after Transferal of the Symbiotic Plasmid pDOA9.Microbes Environ. 2022;37(2):ME22008. doi: 10.1264/jsme2.ME22008. Microbes Environ. 2022. PMID: 35676049 Free PMC article.
-
[Evolution and phylogeny of rhizobia].Rev Latinoam Microbiol. 2005 Jan-Jun;47(1-2):43-60. Rev Latinoam Microbiol. 2005. PMID: 17061545 Review. Spanish.
-
The genetic and biochemical basis for nodulation of legumes by rhizobia.Crit Rev Biotechnol. 1996;16(1):1-51. doi: 10.3109/07388559609146599. Crit Rev Biotechnol. 1996. PMID: 8935908 Review.
Cited by
-
The ever-evolving world of microbes: the current state of microbial taxonomy, genome evolutionary dynamics, and the potential impact on the future of agricultural microbials risk assessment.Front Bioeng Biotechnol. 2025 Jun 23;13:1620652. doi: 10.3389/fbioe.2025.1620652. eCollection 2025. Front Bioeng Biotechnol. 2025. PMID: 40625851 Free PMC article. Review.
-
Integrating multiomics and modern breeding tools for accelerating genetic improvement in Annonas.Funct Integr Genomics. 2025 Jul 12;25(1):155. doi: 10.1007/s10142-025-01653-7. Funct Integr Genomics. 2025. PMID: 40646364 Free PMC article. Review.
References
-
- Bach E., Sant’Anna F. H., Seger G. D., dos S., Passaglia L. M. P. (2022). Pangenome inventory of Burkholderia sensu lato, Burkholderia sensu stricto, and the Burkholderia cepacia complex reveals the uniqueness of Burkholderia catarinensis . Genomics 114, 398–408. doi: 10.1016/j.ygeno.2021.11.011 - DOI - PubMed
LinkOut - more resources
Full Text Sources

