Profiling type I and II interferon responses reveals distinct subgroups of pediatric patients with autoinflammatory disorders
- PMID: 40242151
- PMCID: PMC12002218
- DOI: 10.1016/j.jacig.2025.100450
Profiling type I and II interferon responses reveals distinct subgroups of pediatric patients with autoinflammatory disorders
Abstract
Background: Elevation of type I interferon (IFN-I) is characteristic of a group of diseases known as type I interferonopathies. Several technologies are available to monitor IFN-I, but there is no consensus on their routine use in medical laboratories.
Objective: We aimed to compare the performance of two technologies for this purpose: NanoString, which monitors messenger RNA expression of interferon-stimulated genes (ISGs), and Simoa, which quantifies IFN-α2 protein in an ultrasensitive way. We also designed a NanoString assay to monitor type II ISGs and tested its value to discriminate clinical conditions.
Methods: A total of 196 samples from patients with diseases associated or not with IFN-I pathway activation were analyzed by NanoString and Simoa.
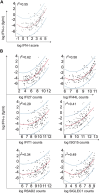
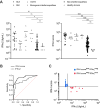
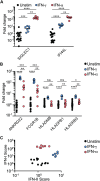
Results: The comparison between NanoString IFN-I score and IFN-α2 Simoa revealed a r 2 coefficient of 0.55. We identified IFI27, IFI44L, and SIGLEC1 as the ISGs most closely related to IFN-α2 concentration. Nineteen samples had a positive IFN-I score but undetectable IFN-α2. These samples were also positive according to IFN-II score, pointing to IFN-II as the primary ISG inducer in corresponding patients. By measuring IFN-I and IFN-II scores in a subset of patients with systemic lupus erythematosus and systemic juvenile idiopathic arthritis, we identified two subgroups of patients in whom IFN-I and IFN-II were dominant.
Conclusion: Both IFN-α2 quantification and NanoString reliably distinguish type I interferonopathies from other diseases. Type I and II interferons induce different transcriptomic signatures in vitro and in vivo, and our results highlight the value of monitoring both IFN-I and IFN-II in interferon-related diseases.
Keywords: IFN signature; Interferonopathy; NanoString; Simoa; juvenile idiopathic arthritis; lupus; type I interferon; type II interferon.
© 2025 The Authors.
Conflict of interest statement
Supported by the 10.13039/501100006451Hospices Civils of Lyon (AO PAM BAP ACP). Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Figures

References
-
- Schoggins J.W. Interferon-stimulated genes: what do they all do? Annu Rev Virol. 2019;6:567–584. - PubMed
LinkOut - more resources
Full Text Sources

