Serum proteomics of adults with acute liver failure provides mechanistic insights and attractive prognostic biomarkers
- PMID: 40242314
- PMCID: PMC11998117
- DOI: 10.1016/j.jhepr.2025.101338
Serum proteomics of adults with acute liver failure provides mechanistic insights and attractive prognostic biomarkers
Abstract
Background & aims: Acute liver failure (ALF) is defined as rapid onset coagulopathy and encephalopathy in patients without a prior history of liver disease. We performed untargeted and targeted serum proteomics to delineate processes occurring in adult patients with ALF and to identify potential biomarkers.
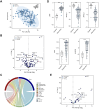
Methods: Sera of 319 adult patients with ALF (∼50% acetaminophen [APAP]-related cases) were randomly selected from admission samples of the multicenter USA Acute Liver Failure Study Group consortium and subdivided into discovery/validation cohorts. They were analyzed using untargeted proteomics with mass spectroscopy and a serum cytokine profiling and compared with 30 healthy controls. The primary clinical outcome was 21-day transplant-free survival. Single-cell RNAseq data mapped biomarkers to cells of origin; functional enrichment analysis provided mechanistic insights. Novel prognostic scores were compared with the model for end-stage liver disease and ALFSG prognostic index scores.
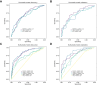
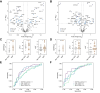
Results: In the discovery cohort, 117 proteins differed between patients with ALF and healthy controls. There were 167 proteins associated with APAP-related ALF, with the majority being hepatocyte-derived. Three hepatocellular proteins (ALDOB, CAT, and PIGR) robustly and reproducibly discriminated APAP from non-APAP cases (AUROCs ∼0.9). In the discovery cohort, 37 proteins were related to 21-day outcome. The key processes associated with survival were acute-phase response and hepatocyte nuclear factor 1α signaling. SERPINA1 and LRG1 were the best individual discriminators of 21-day transplant-free survival in both cohorts. Two models of blood-based proteomic biomarkers outperformed the model for end-stage liver disease and ALFSG prognostic index and were reproduced in the validation cohort (AUROCs 0.83-0.86) for 21-day transplant-free survival.
Conclusions: Proteomics and cytokine profiling identified new, reproducible biomarkers associated with APAP etiology and 21-day outcome. These biomarkers may improve prognostication and understanding of the etiopathogenesis of ALF but need to be independently validated.
Impact and implications: Acute liver failure (ALF) is a sudden, and severe condition associated with high fatality. More sensitive and specific prognostic scores are urgently needed to facilitate decision-making regarding liver transplantation in patients with ALF. Our proteomic analysis uncovered marked differences between acetaminophen and non-acetaminophen-related ALF. The identification of routinely measurable biomarkers that are associated with 21-day transplant-free survival and the derivation of novel prognostic scores may facilitate clinical management as well as decisions for/against liver transplantation. Further studies are needed to quantify less abundant proteins. Although we used two cohorts, our findings still need to be independently and prospectively validated.
Keywords: ALF subtyping; Acetaminophen; Acute liver injury; Proteomic profiling.
© 2025 The Author(s).
Conflict of interest statement
PS reports receiving grant support and lecture fees from Grifols and CSL Behring, grant support and advisory board fees from Arrowhead Pharmaceuticals, grant support from Vertex Pharmaceuticals, advisory board fees from Dicerna Pharmaceuticals and Ono Pharmaceuticals, and lecture fees from Alnylam Pharmaceuticals. RJF has received research support from Kezar Pharmaceuticals, Takeda Pharmaceuticals, and the NIH (ALFSG and DILIN). WML consults for Genentech, SeaGen, GSK, and Veristat and receives research support from Gilead, Alexion, Vivet, Camurus, and Lipocine, none related to the current article. BE was supported by the PRACTIS – Clinician Scientist program of Hannover Medical School, funded by the German Research Foundation (DFG, ME 3696/3). All other authors report no conflicts of interest. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures



References
-
- European Association for the Study of the Liver EASL Clinical Practical Guidelines on the management of acute (fulminant) liver failure. J Hepatol. 2017;66:1047–1081. - PubMed
-
- McPhail M.J.W., Farne H., Senvar N., et al. Ability of King’s College Criteria and model for end-stage liver disease scores to predict mortality of patients with acute liver failure: a meta-analysis. Clin Gastroenterol Hepatol. 2016;14:516–525.e5. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

